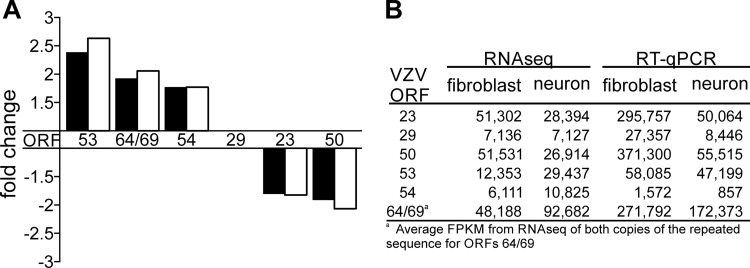
FIG 4.
Validation of RNA-seq by RT-qPCR. RNA used in RNA-seq analysis was reverse transcribed with oligo(dT), and primers and cDNA were analyzed by RT-qPCR. Primer/probe sets were designed for five VZV ORFS that exceeded the ±1.70-fold cutoff; three genes (ORFs 53, 64/69, and 54) were transcribed more in neurons, and two genes (ORFs 23 and 50) were transcribed less in neurons. Each RT-qPCR mixture contained a primer/probe set for ORF 29 for normalization. (A) Fold changes (from neurons to fibroblasts) in the levels of transcription of the six ORFs from RNA-seq analysis (black bars) or by RT-qPCR after normalization to ORF 29 (white bars). (B) Raw data from RNA-seq (FPKMs) and RT-qPCR (copy numbers) used to construct the graph in panel A.