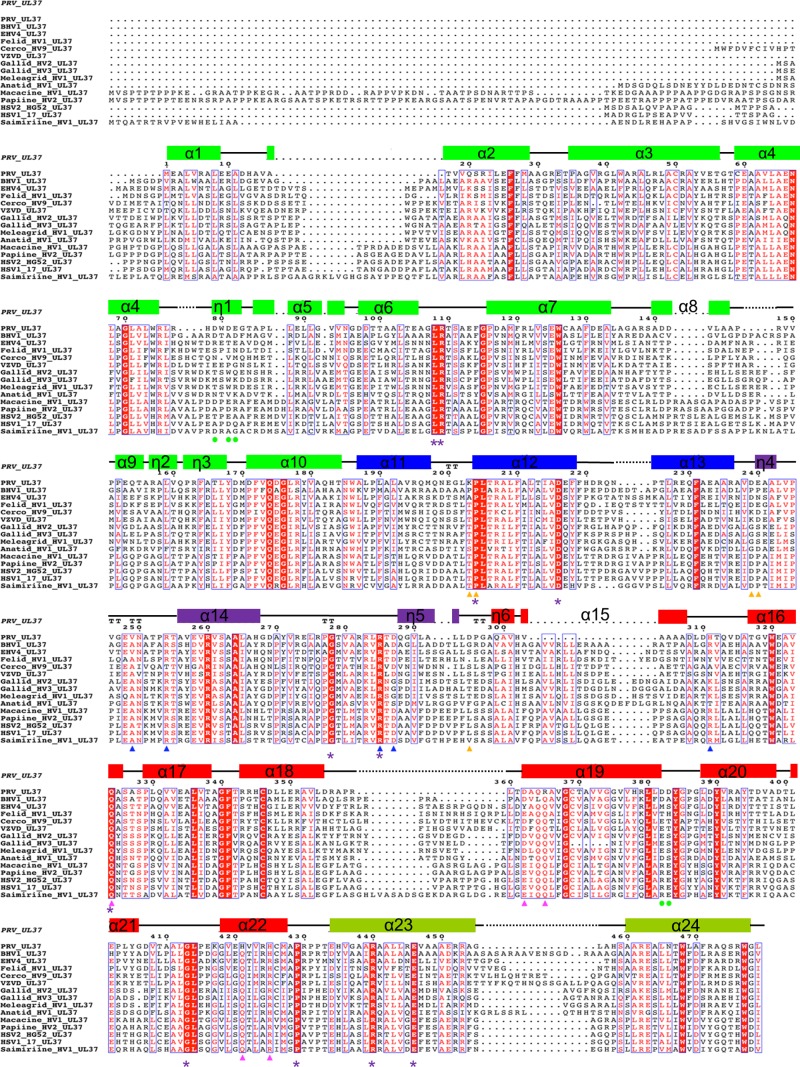
FIG 4.
Multiple-sequence alignment of UL37 homologs from 15 alphaherpesviruses. Only the alignment of residues corresponding to residues 1 to 479 of PRV UL37 is shown. Helices are indicated by rectangular boxes labeled and colored by domain with colors similar to those used in Fig. 1C. Identical residues are boxed in red, and those exposed on the surface of UL37N are marked by purple asterisks. Residues involved in binding calcium are marked by green circles. All residues specified for mutagenesis are highlighted by triangles (blue, R1; pink, R2; orange, R3) using the same color scheme used in Fig. 5C. PRV, suid herpesvirus strain Kaplan; BHV1, bovine herpesvirus 1; EHV1, equine herpesvirus 1; Felid_HV1, felid herpesvirus 1; Cerco_HV9, cercopithecine herpesvirus 9; VZVD, varicella-zoster virus serotype D; Gallid_HV2, gallid herpesvirus 2; Gallid_HV3, gallid herpesvirus 3; Meleagrid_HV1, meleagrid herpesvirus 1; Anatid_HV1, anatid herpesvirus 1; Macacine_HV1, macacine herpesvirus 1; Papiine_HV2, papiine herpesvirus 2; HSV2_HG52, herpes simplex virus 2 strain HG52; HSV1_17, herpes simplex virus1 strain 17; Saimiriine_HV1, saimiriine herpesvirus 1. PRV_UL37 (in italics) represents the secondary structure derived from the crystal structure of PRV UL37.