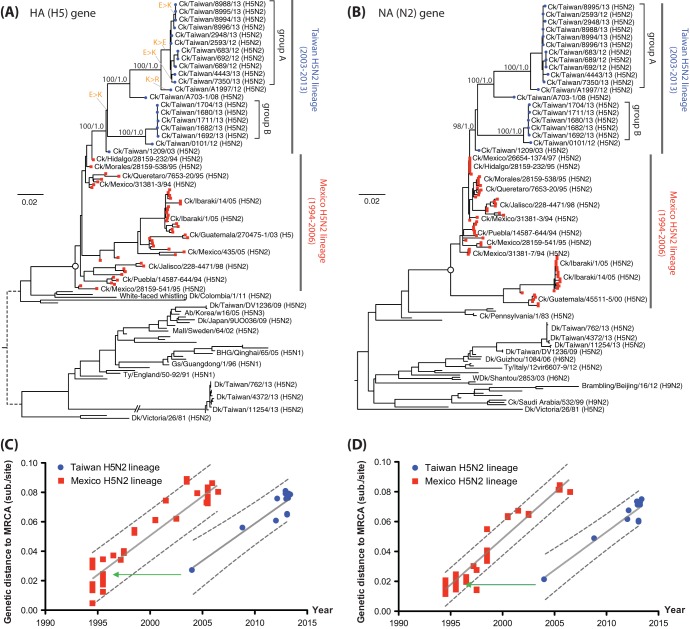
FIG 2.
Phylogenies of the H5 and N2 genes. (A and B) Maximum-likelihood phylogenies of H5 and N2 genes showing the chicken H5N2 viruses from Taiwan. Bootstrap support and Bayesian posterior clade probabilities of selected lineages are shown. Taxa from the H5N2 outbreak in Mexico and the related viruses found in Japan are indicated. The most recent common ancestor (MRCA) of the Mexican-origin H5N2 lineage is indicated by the open circles in the trees. Changes of the amino acid at position -3 of HA1 (Table 3) are indicated in orange and a connecting thin bar. In the HA tree, the long branch leading to the Taiwan H5N2 duck viruses reported in this study has a length of 0.249 substitutions/site. (C and D) Linear regression plots of genetic distances from the MRCA of the Mexican-origin H5N2 lineage against isolation time of the taxa. The estimate and 95% confidence intervals for the evolutionary rate are shown by solid and dashed lines, respectively. Green arrows indicate the isolation dates of Mexican viruses that have similar genetic distances to the MRCA as the 2003 isolate from Taiwan.