FIG 3.
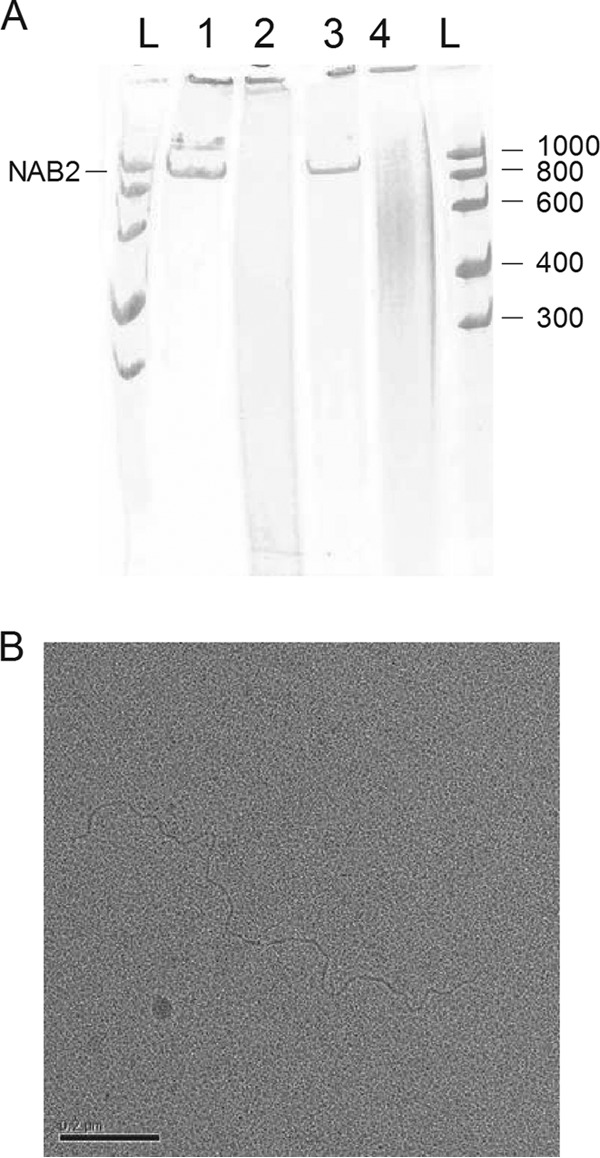
NAB2 configuration. (A) NAB2 was heat denatured in water and renatured in high-concentration MgCl2 buffer. Then, NAB2 was digested with the dsRNA-specific RNase III and RNase V1 and single-strand RNA-specific RNase T2. Urea and dyes were added, and the samples were run on a 6% polyacrylamide-urea slab gel under denaturing conditions. Lane 1, NAB2; lane 2, NAB2 digested with RNase III; lane 3, NAB2 digested with RNase T2; lane 4, NAB2 digested with RNase V1; lanes L, RNA ladder (numbers on the left right are in bases). NAB2 is sensitive to the two dsRNA-specific RNases. (B) Electron microscopy analysis of NAB2. Platinum-shadowed nucleic acid molecules were observed in a transmission electron microscope. Analyses revealed a linear molecule without bulges or forks.
