FIG 1.
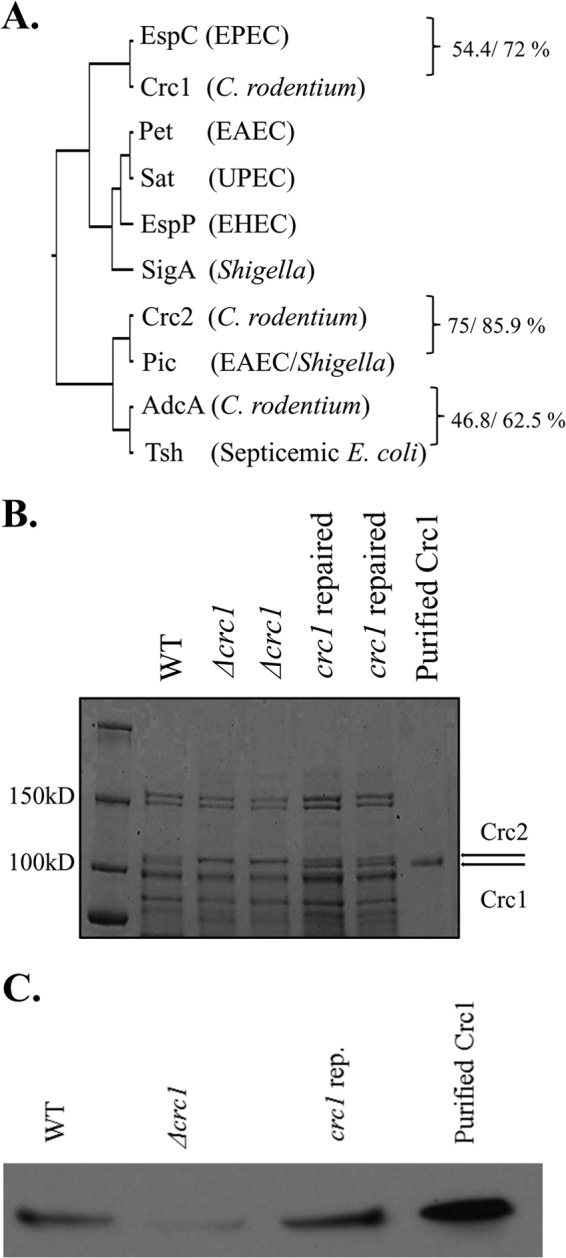
(A) Phylogenetic analysis of C. rodentium SPATEs. Alignment of sequences was built with ClustalW, and the phylogenetic tree was constructed using UPGMA (unweighted pair group method using arithmetic mean). C. rodentium SPATEs and their closest homologs were aligned using the BLOSUM50 alignment matrix, and identity/similarity was calculated using the Matrix Global Alignment Tool. (B) Secretion of Crc1 and Crc2 into the culture supernatant by C. rodentium ICC168. Supernatants from C. rodentium WT, Δcrc1, and crc1-repaired strain bacterial cultures were collected. The supernatant was concentrated 1,000-fold using Amicon filters with a 70-kDa cutoff size. The supernatants were analyzed by SDS-PAGE. Crc2 (predicted molecular mass, 109.3 kDa) and Crc1 (predicted molecular mass, 104.9 kDa) appeared above the 100-kDa band of the molecular mass ladder (shown by arrows). Crc1 purification is illustrated in Fig. S1 in the supplemental material. (C) Confirmation of Crc1 deletion. Supernatants from C. rodentium WT, Δcrc1, and crc1-repaired strain bacterial cultures were collected. The supernatant was concentrated 1,000-fold using Amicon filters with a 70-kDa cutoff size. The supernatants and the purified Crc1 toxin were analyzed by immunoblotting using anti-Crc1 rabbit antibody.
