Figure 3. Architecture of rearrangements between the WI strain and the wild UCB strain.
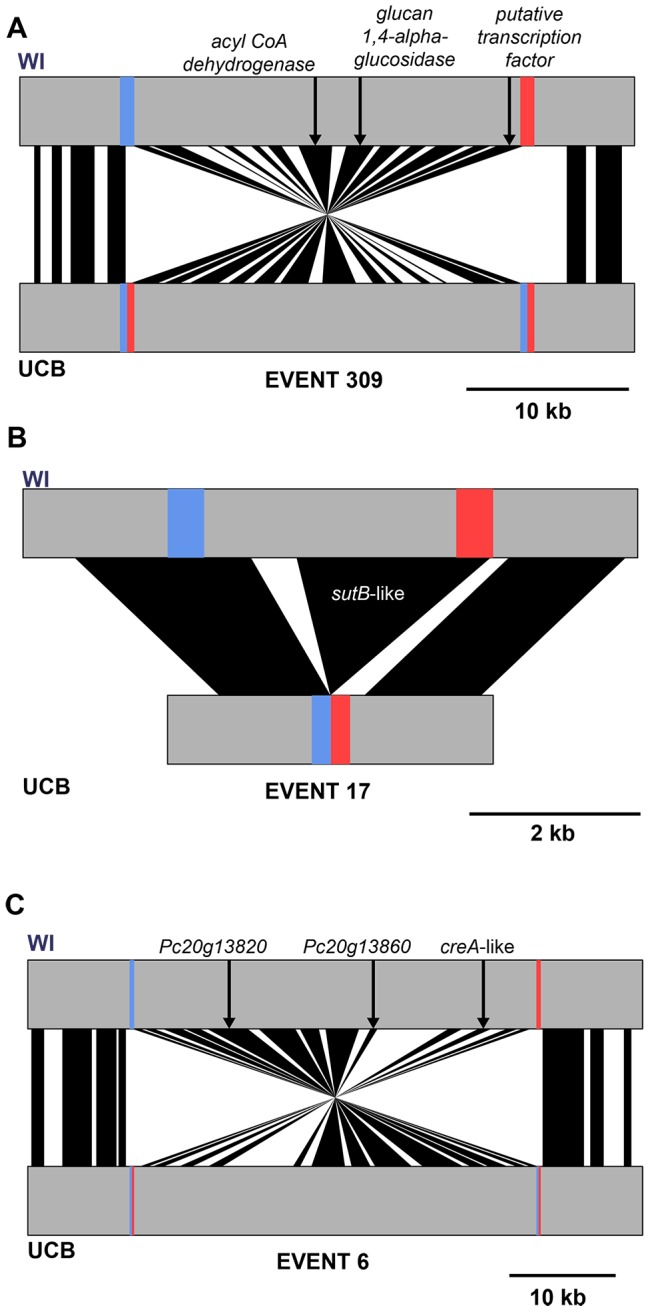
Rearrangement events containing genes with potential roles in penicillin biosynthesis are shown with their predicted gene contents (black spans). Flanking blocs of aberrantly mapping reads are marked in blue and red and penicillin-related genes are denoted by arrows. For those genes with known function, the gene name is listed, otherwise the gene ID is given. (A) Event 309 is an inversion and contains the predicted glucan 1,4-alpha-glucosidase Pc13g11940 and the gene of unknown function Pc13g11930, both of which show elevated expression in high penicillin G producing strains [16], [20] as well as Pc13g11930, which is predicted to localize to the microbody where penicillin biosynthesis takes place. (B) Event 17 is an insertion of Pc12g01540, a gene with strong similarity to the sulfate transporter sutB, which also shows elevated expression in high penicillin-producing strains and may be involved in biosynthesis of amino acid β-lactam precursors [16], [20]. (C) Event 6 is an inversion containing three genes that are repressed in high penicillin-producing strains: Pc20g13820, Pc20g13860, and Pc20g13880 [16], the latter of which is a homolog of the Aspergillus niger creA regulator of β-lactam biosynthesis.
