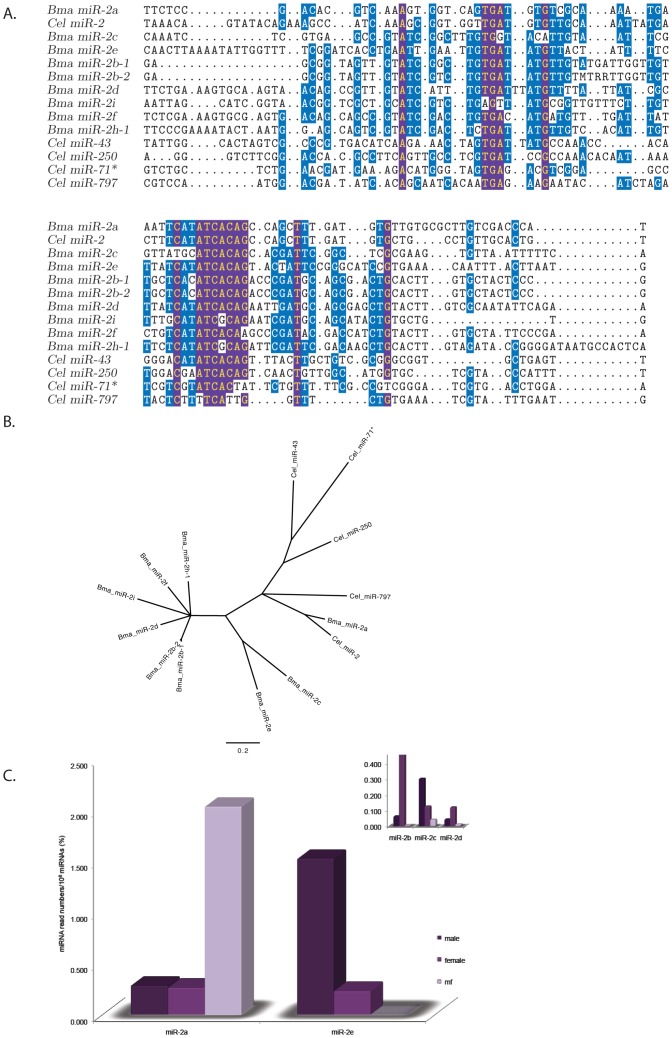
Figure 5. Alignment and Expression Analysis of the B. malayi miR-2 family.
(A) Hairpin alignment of the miR-2 paralogues from B. malayi and C.elegans. Nucleotides conserved in ≥80% of the sequences making up the alignment are highlighted yellow in purple boxes. Nucleotides conserved in 50% of the sequences making up the alignment are highlighted white in blue boxes. The Bma-miR-2g hairpin is not included in the alignment because the mature miR is located on the 5′ arm not the 3′ arm, the location of the other mature miR-2 paralogues. (B). An un-rooted tree calculated from the alignment in A. The 0.2 bar represents nucleotide substitutions per site. (C). Bar graph depicting the expression levels of the prevalent B. malayi miR-2 paralogues (Tables 4, S2 and S3) in the adult male, female and mf CIP libraries. Bma-miR-2b, -2c and -2d are shown in the inset.