Fig. 1.
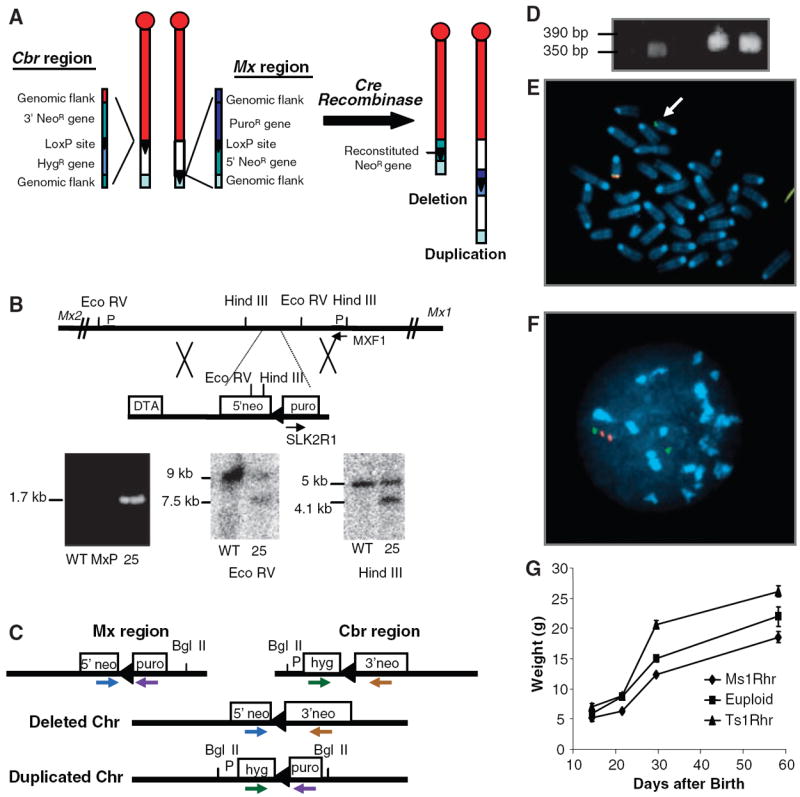
Construction of a duplication or deletion of the MMU16 region orthologous to the DSCR. (A) LoxP sites were targeted to asymmetrical positions on MMU16 at Cbr1 (13) and adjacent to Mx2. Each targeting vector contained a LoxP site (triangle), a selectable antibiotic resistance gene (hyg or puro), and half of the neomycin resistance gene (5′ or 3′ neo). (B) The Mx-Lox vector produced a 1.7-kb PCR product from the targeted allele in line MxP25. Wild-type (WT) and targeted alleles produced 9.0- and 7.5-kb restriction fragments with EcoRV and 5.0- and 4.1-kb fragments with HindIII. Arrows identify PCR primers; P designates probes. (C) PCR primers used to screen vector sequences for recombination after Cre-mediated translocation. (D) PCR products from neor ES lines produced the 350– and 390–base pair (bp) fragments expected for deletion and duplication, respectively. (E) Metaphase FISH with one bacterial artificial chromosome (BAC) that maps to the DSCR (red) and a second BAC proximal to it (green) shows one chromosome with a single green signal (arrow) and a second with green and red plus a yellow signal, indicating overlap. (F) Interphase FISH shows a green signal by itself representing the deleted MMU16 and a green signal with two adjacent red signals representing the duplication. (G) Body weights of Ts1Rhr mice are significantly larger than controls. Standard errors (bars) are indicated.
