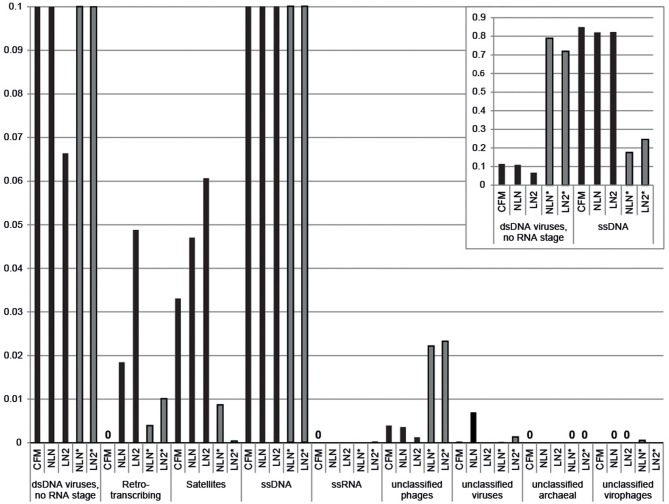
Figure 3.
Annotation of Pocillopora damicornis DNA virome metagenomic sequences with a match to viral sequences for each method and treatment. Best matches to major viral groups were examined through Metavir, which uses tBLASTx algorithm against the NCBI viral Refseq genomes with a bit-score of 50 (data sets are publically available at http://metavir-meb.univ-bpclermont.fr, project “Coral virus—generating metagenomes”). Total numbers (per cent) of sequences for each viral group are indicated in Table 1. All samples were amplified using a RepliG® kit except for samples NLN* and LN2*, which underwent RP-SISPA amplification.