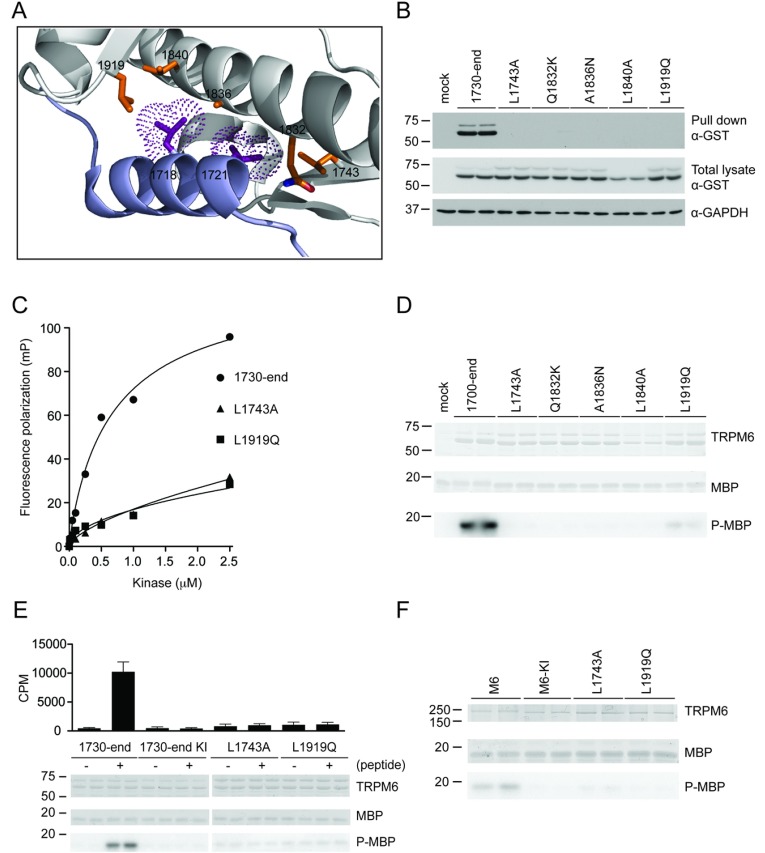
Figure 3. Analysis of the dimerization pocket residues.
(A) Structural model of TRPM6 based on TRPM7 α-kinase depicting the binding of Leu1718 and Leu1721 to other residues in the α-kinase domain. (B) GST-tagged TRPM6-(1730–end) and the indicated mutants were expressed in HEK-293 cells. Subsequently, cell lysate was subjected to a peptide pull-down, as described in the Materials and methods section, using wild-type dimerization motif peptide. The samples were analysed by immunoblotting with anti-GST antibody (top panel). Total cell extracts were immunoblotted with anti-GST antibody (middle panel), using GAPDH expression as loading control (bottom panel). (C) Fluorescence polarization analysis of the dimerization motif peptide with E. coli purified GST-tagged TRPM6-(1730–end) and indicated mutants. (D) Immunoprecipitation of GST-tagged TRPM6-(1700–end) and indicated mutants from HEK-293 cell lysate was followed by a kinase assay. Proteins were separated by SDS/PAGE, stained with Coomassie Blue (top and middle panels) and MBP phosphorylation was detected by autoradiography (bottom panel). (E) GST-tagged TRPM6-(1730–end), both wild-type and kinase-inactive (KI), and L1743A and L1919Q mutants were expressed in HEK-293 cells, followed by immunoprecipitation and then subjected to a kinase assay without (−) or with (+) 30 μM dimerization motif peptide. Proteins were separated by SDS/PAGE, stained with Coomassie Blue (top and middle gels) and MBP phosphorylation was detected by autoradiography (bottom gel). Incorporated radioactive counts are depicted as a histogram. (F) Immunoprecipitated FLAG-tagged TRPM6 and the indicated mutants were subjected to a kinase assay using MBP as substrate. Proteins were separated by SDS/PAGE, stained with Coomassie Blue (top and middle panels) and MBP phosphorylation was detected by autoradiography (bottom panel). Molecular masses are indicated in kDa.