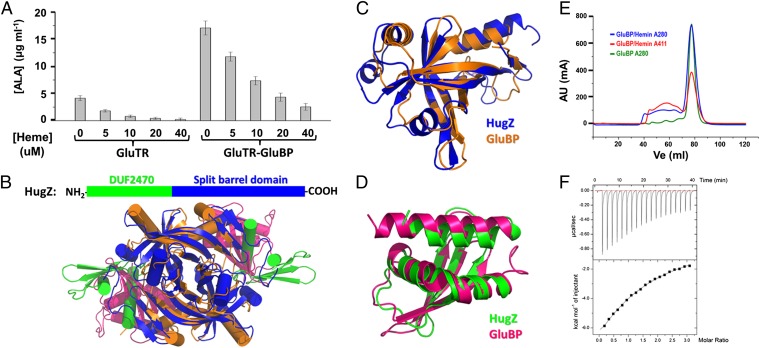
Fig. 5.
GluBP belongs to a heme-binding protein family. (A) Heme represses ALA synthesis in a concentration-dependent manner. Data are presented as the mean ± SEM of six independent experiments. (B) Ribbon representation showing the overall structures of GluBP and HugZ. (C) Superposition of the FMN-binding split barrel domains. (D) Superposition of the DUF2470 domains. (E) Elution profiles of GluBP in the absence (green line) and presence (blue line) of hemin. After incubation with a twofold excess of hemin, GluBP sample was subjected to Superdex 200 column. The red line corresponds to absorbance at 411 nm. (F) ITC titration of GluBP with hemin. The fitted parameters for a single site binding model are as follows: n = 1.08 ± 0.14, Kd = 0.20 ± 0.02 mM, ΔH = −21.6 ± 3.5 kcal/mol, and ΔS = −56.7 cal/mol/K.