Fig. 1.
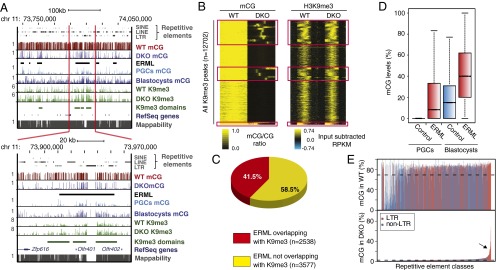
Genome-wide profile of mCG in wild-type and Dnmt3a,3b double knockout (DKO) mESCs. (A, Upper) University of California Santa Cruz (UCSC) genome browser screen capture showing the DNA methylation profiles for WT and DKO mESCs, mouse E13.5 male primordial germ cells (PGCs) (17), and blastocyst (16) across a region on chromosome 11. ERML are denoted with black bars. H3K9me3 ChIP-seq reads from WT and DKO are also included. H3K9me3-enriched domains in WT are called “ChromaBlocks.” Genes and repetitive elements as annotated by RefSeq and RepeatMasker, respectively, are shown. MethylC-seq reads from WT and DKO are mapped, with 100-bp binned data showing mCG ratios with bar heights between 0 and 1. H3K9me3 profiles are shown as reads per kilobase pair per million reads (RPKM). Mappability of reference genome from the ENCODE project for 50-bp segments is shown with the score between 0 and 1. (Lower) Screen capture of a narrowed segment within the same region as shown above. (B) Heat map generated by k-means clustering shows the H3K9me3 enrichment (Right) at all identified peaks and mCG (Left) in WT and DKO mESCs. Red boxes outline regions retaining mCG in the DKO cells, which we term ERML. These sites are enriched for strongest K9me3 peaks. (C) Pie chart summarizing the overlap between regions enriched for H3K9me3 in both WT and DKO mESCs and ERML. (D) Boxplot showing the difference in mCG levels at ERML and control shuffled regions in PGCs and blastocysts. (E) Barplot of mCG ratios of mappable LTR (red) and non-LTR (blue) repeats in WT (Upper) and DKO (Lower) mESCs. Arrow indicates families that retain higher levels of mCG.
