Fig. 2.
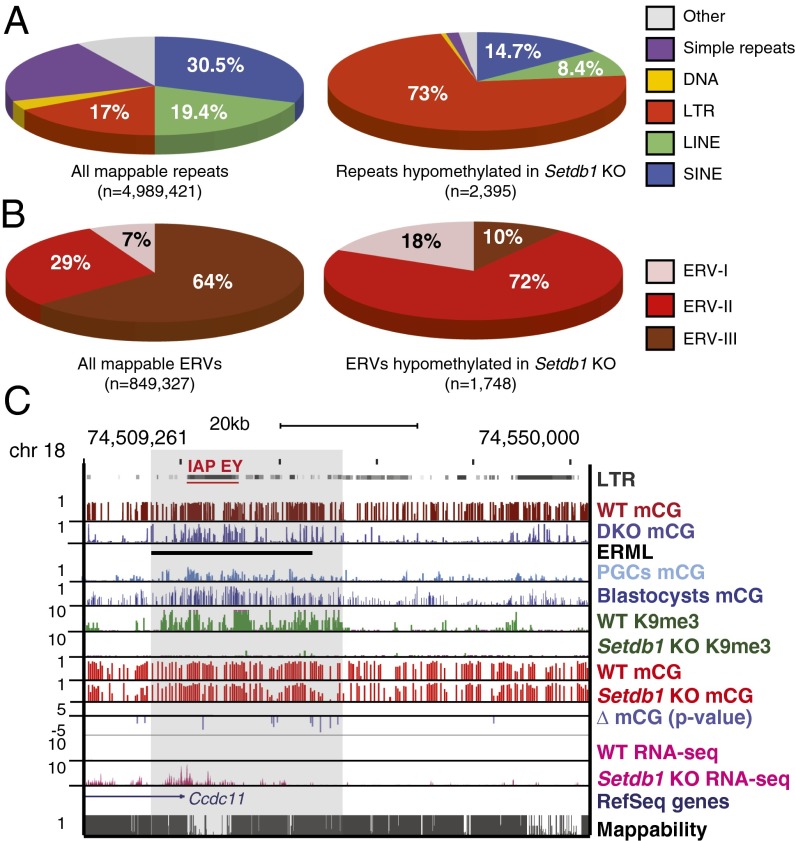
Subsets of endogenous retroviruses and ERML show significant mCG reduction in Setdb1 KO mESCs. (A) Pie charts demonstrating the distribution of repetitive element classes among all mappable repeats (n = 4,989,421) in the mouse mm9 reference genome [short interspersed nuclear elements (SINEs) = 30.5%, long interspersed nuclear elements (LINEs) = 19.4%, simple repeats = 21.3%, LTR = 17%, DNA transposons = 3.1%, and others = 8.4%] (Left) and significantly hypomethylated elements (n = 2,395) in Setdb1 KO mESCs (SINE = 14.7%, LINE = 8.4%, simple repeats = 1.6%, LTR = 73%, DNA transposons = 0.5%, and others = 1.7%) (Right). (B) Pie charts demonstrating the distribution of the three ERV classes among all mappable ERVs (n = 849,327) (Left) and significantly hypomethylated elements (n = 1,748) in Setdb1 KO mESCs. (C) Screen capture illustrating the overlap of ERML and an IAP-EY endogenous retroviral element. MethlyC-seq reads from WT (predeletion) and Setdb1 KO mESCs are mapped and divided into 100-bp bins, showing mCG ratios with bar heights between 0 and 1. Log P values (Fisher’s exact test) are shown for bins with significant difference in mCG ratio between WT and Setdb1 KO (P value cutoff of 0.001 or log = −3). RNA-seq (20) confirms substantial reactivation of this element in Setdb1 KO mESCs. Mappability or uniqueness of reference genome from the ENCODE project for 50-bp segments is shown with the score between 0 and 1.