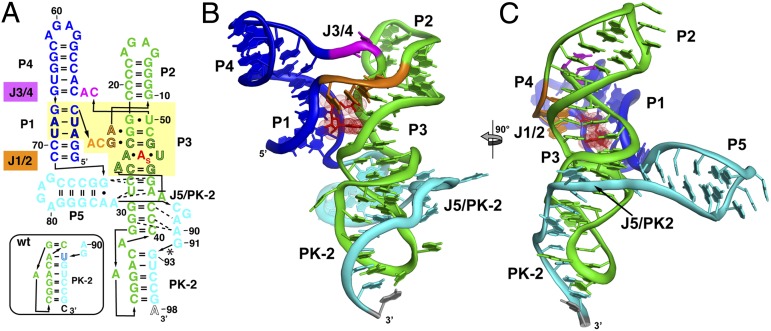
Fig. 2.
Crystal structure of the env8 SAM-I/IV riboswitch aptamer. (A) Sequence of the crystallized RNA [env8[(∆U92); site of deletion denoted by asterisk] drawn to reflect the tertiary architecture of the RNA. The yellow box represents sequence (black outlined letters) and/or structural elements that are nearly universally conserved in the SAM clan, with the site of binding of the adenosyl moiety of SAM represented as “A”s. Coloring of the RNA is used to highlight the P1/P4 coaxial stack (blue), the P2/P3 coaxial stack (green), the joining regions between the stacks (orange and magenta), and the PK-2 subdomain (cyan). The box represents the most probable secondary structure of PK-2 for the wild-type sequence that was used for modeling. (B) Cartoon representation of the global architecture of the RNA, using the same coloring scheme as in A. (C) 90° clockwise rotation perspective of the structure.