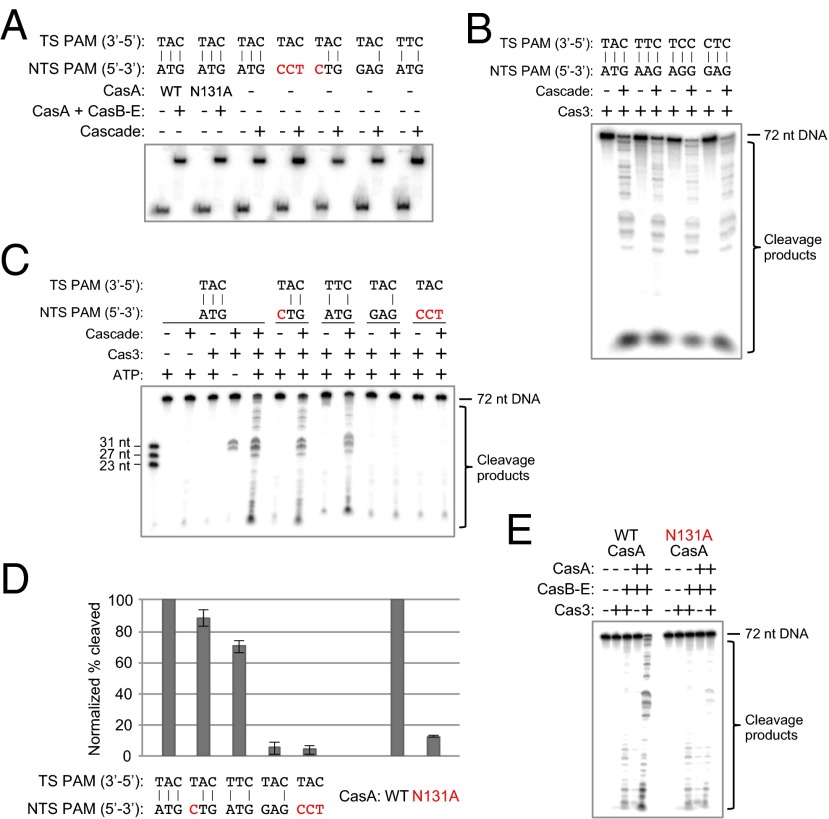
Fig. 3.
Target cleavage by Cas3 requires base pairing within the PAM sequence and integrity of the PAM-binding loop in CasA. (A) Native gel electrophoretic mobility-shift assay showing that all dsDNA substrates tested for Cas3-mediated degradation are fully bound by Cascade under the conditions used in the cleavage assays. Red text indicates a nucleotide mutation that changes the PAM to a sequence other than one of the four functional motifs. Cascade (1 µM) was incubated with 0.1–0.5 nM [32P]dsDNA at 37 °C for 30 min before gel-shift analysis by 10% native PAGE. NTS, non-target strand; TS, target strand. (B) All four PAM sequences that function in vivo also permit target cleavage in vitro. DNA cleavage reactions were performed using target dsDNAs with different PAM sequences at a final concentration of 1 nM in 1× reaction buffer. In the presence of 2 mM ATP, 1 μM targeting Cascade, and 500 nM MBP-tagged Cas3, all four targets are cleaved after incubation at 37 °C for 30 min. (C) PAM base-pair mismatches prevent efficient target cleavage by Cas3. Targeting Cascade was prebound to each dsDNA before addition of ATP and MBP-Cas3. (D) Quantified cleavage percentages for each condition in the presence of Cas3 and ATP are the average of three (E) or four (C) independent replicates; error bars represent ±1 SD. All values have been normalized to the WT cleavage efficiency. (E) Denaturing polyacrylamide gel depicting the cleavage defect caused by mutation of Asn131 in the L1 loop of CasA. Wild-type or N131A CasA (1 μM) was added back to 1 μM Strep-tag II–tagged CasB–E to reconstitute the Cascade complex and prebound to a 1 nM dsDNA target with a functional PAM before addition of ATP and MBP-Cas3.