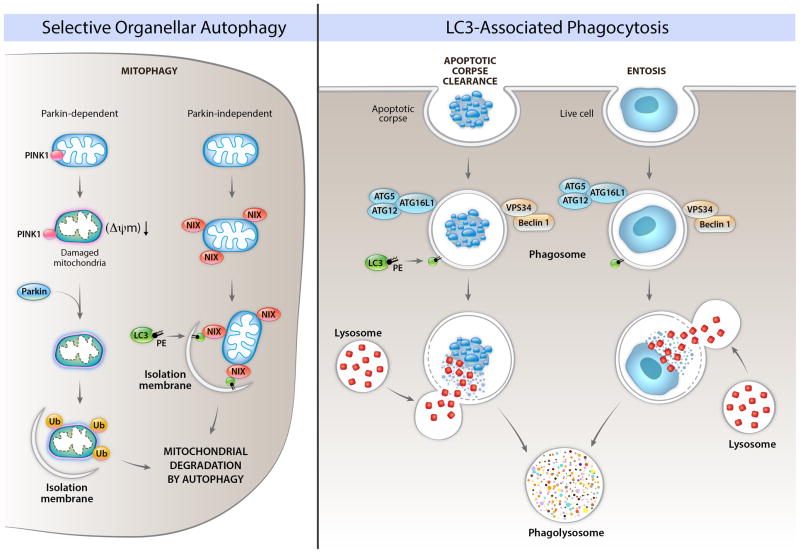
Figure 2. Roles of autophagy proteins in the removal of unwanted organelles and in the removal of cells.
The left panel shows Parkin-dependent and Parkin-independent mechanisms involved in the selective degradation of mitochondria by autophagy (mitophagy). In Parkin-dependent mitophagy, mitochondrial damage and loss of mitochondrial membrane potential (ΔΨm) leads to localization of the kinase, PINK1, on the cytoplasmic surface of the mitochondria, resulting in recruitment of the E3 ubiquitin ligase, Parkin, to the mitochondria, followed by the ubiquitination of mitochondrial proteins, and the formation of an isolation membrane that surrounds the damaged mitochondria. In Parkin-independent mitophagy, protein such as Nix (shown in figure), BNIP3, and FUNDC1 (not shown in figure) bind to LC3. Other autophagy proteins may be involved in Parkin-dependent and Parkin-independent mitophagy (discussed in text). The precise details of how an isolation membrane is formed around specific mitochondria earmarked for degradation are unclear. Other damaged/unwanted organelles such as ER, peroxisomes, and lipid droplets can also be degraded by selective autophagy; the molecular mechanisms of these forms of selective autophagy are not well understood in mammalian cells. The right panel depicts roles of LC3-associated phagocytosis (LAP) of apoptotic corpses and of live cells (entosis). In LAP, components of the autophagy initiation complex (Beclin 1, VPS34) are recruited to the phagosome, which leads to recruitment of LC3-PE, and facilitation of phagolyosomal fusion. This process requires other components of the elongation machinery, but – in contrast to general autophagy or selective autophagy – proceeds independently of the ULK pre-initiation complex.