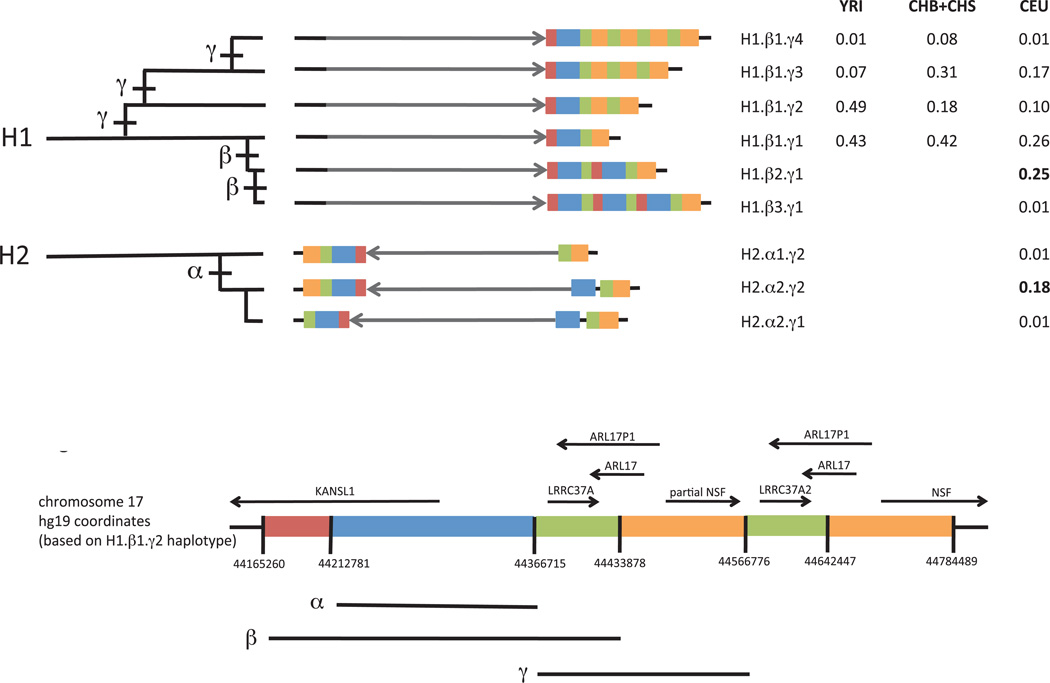
Figure 2.
Structural forms of the human 17q21.31 locus and their frequencies in populations. Each haplotype is represented in a simplified form to highlight major structural differences. The schematic at bottom indicates which genomic segment is represented by each color; detailed schematics with physical coordinates are available in Supplementary Material. The grey arrow indicates orientation of the unique inverted region within 17q21.31. Duplications of a 150-kb genomic segment (blue) containing the 5’ exons of the KANSL1 gene appear to have arisen on both the H1 and H2 forms of the 17q21.31 inversion polymorphism and reached high allele frequency in West Eurasian populations. The H1-polymorphic duplication β (red, blue, green) is longer than the H2-polymorphic duplication α (blue). A third duplication polymorphism γ (orange, green) affecting the NSF gene also varies in copy number. These structural polymorphisms segregate as the nine common haplotypes shown. The H2 inversion form shows structural diversity that was heretofore unappreciated, including a simpler, less common structural form (H2.α1) that may be the ancestral H2 structure. The table to the right lists allele frequencies for the nine structural haplotypes in different populations. CEU: Utah residents with Northern and West European ancestry. CHB: Han Chinese in Beijing. CHS: Han Chinese South. YRI: Yoruba in Ibadan, Nigeria. Genotype and allele frequencies in 12 populations are available as Supplementary Tables 2–9. Most of these haplotypes correspond one-to-one to haplotypes identified in the contemporaneous work by Steinberg et al.34: H1.β1.γ1 corresponds to H1.1; H1.β1.γ2 to H1.2 ; H1.β1.γ3 to H1.3; H1.β2.γ1 to H1D; H1.β3.γ1 to H1D.3; H2.α1.γ1 to H2.1; H2.α1.γ2 to H2.2; and H2.α2.γ2 to H2D.