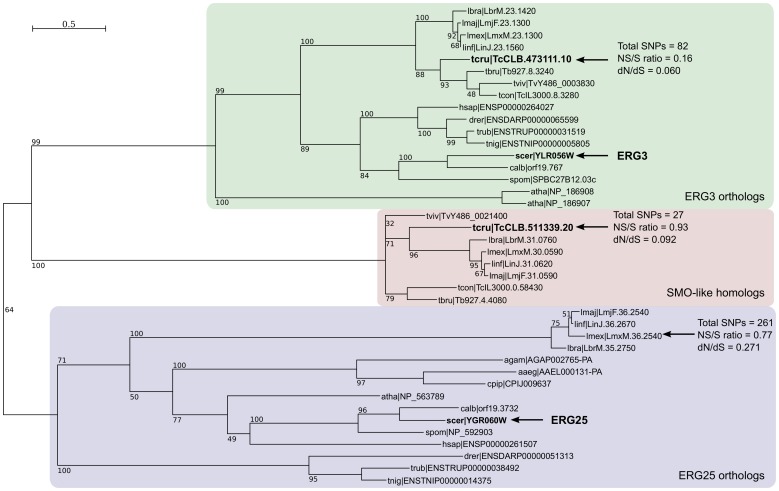
Figure 1. Phylogenetic tree of selected ERG3/ERG25 orthologs.
Seleced orthologs from kinetoplastids, plants, vertebrates, invertebrates and fungi were aligned with t_coffee. A phylogenetic reconstruction was calculated using PhyML (LG model, bootstrap resampling with N = 1000). For clarity, highly similar genes/alleles were not included in the final tree. Organism abbreviations are: lbra = L. braziliensis; lmex = L. mexicana; linf = L. infantum; lmaj = L. major; tcru = T. cruzi; tviv = T. vivax; tcon = T. congolense; tbru = T. brucei; agam = Anopheles gambiae; aaeg = Aedes aegypti; cpip = Culex pipiens; hsap = Homo sapiens; calb = Candida albicans; scer = S. cerevisiae; spom = Schizosaccharomyces pombe; atha = A. thaliana; drer = Danio rerio; trub = Takifugu rubripes; tnig = Tetraodon nigroviridis.