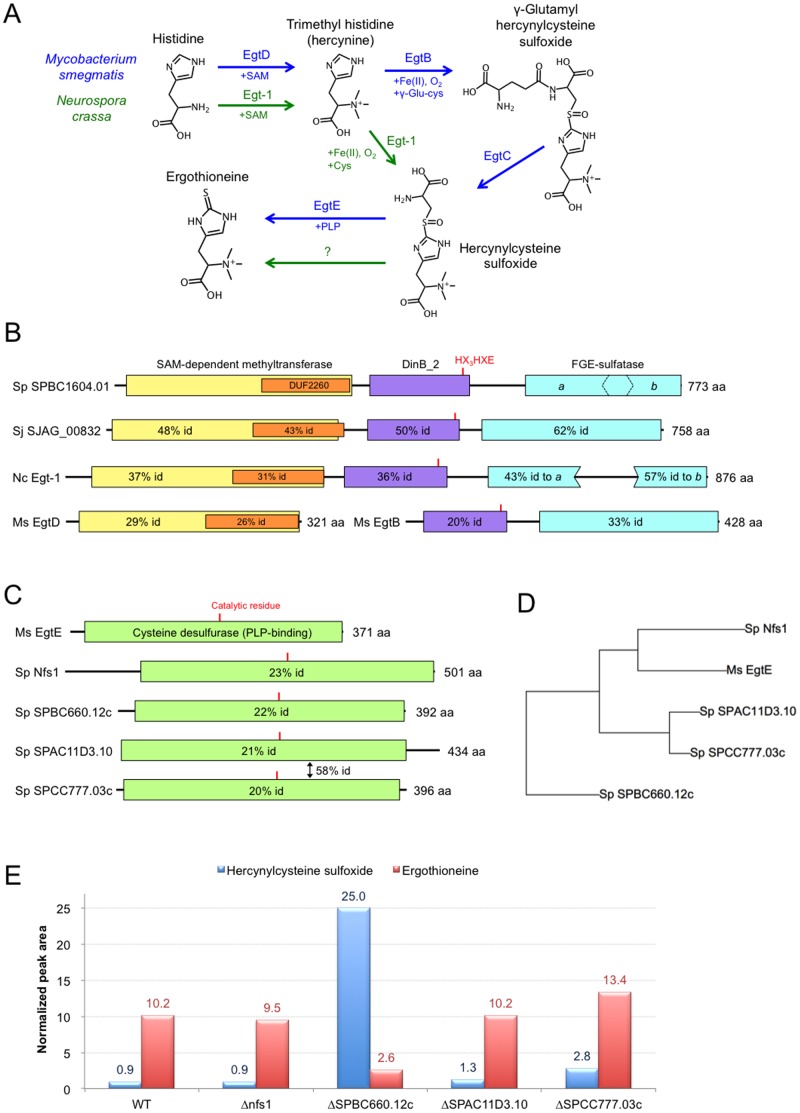
Figure 1. Characterization of EGT biosynthesis in S. pombe.
A. Previously published EGT biosynthesis pathways in M. smegmatis and N. crassa. The Egt-1 protein in N. crassa is a fusion enzyme that catalyzes two different reactions. B. Comparison of the conserved domain structure of S. pombe protein SPBC1604.01 (designated Egt1 in this manuscript) with S. japonicus SJAG_00832 and EGT biosynthesis proteins in N. crassa (Egt-1) and M. smegmatis (EgtD and EgtB). All proteins are composed of: 1. an S-adenosyl-L-methionone (SAM)-dependent methyltransferase domain, including DUF2260, a domain of unknown function; 2. an uncharacterized DinB_2 domain, including an iron-binding motif HX3HXE; and 3. a formylglycine generating enzyme (FGE)-sulfatase domain. Percentage identity (% id) of amino acid sequences is indicated in comparison to the corresponding sequence in S. pombe. C. Comparison of conserved domain structure of M. smegmatis protein EgtE with its four putative homologs in S. pombe. All proteins contain a single pyridoxal phosphate (PLP)-binding cysteine desulfurase domain. The conserved catalytic residues (PLP binding sites) are indicated by red lines. Percentage identity (% id) of the amino acid sequences is indicated in comparison to the corresponding sequence in M. smegmatis. D. Phylogenetic tree visualizing the similarity of amino acid sequences of M. smegmatis EgtE and its putative homologs in S. pombe. E. Normalized peak areas of hercynylcysteine sulfoxide and EGT obtained by metabolomic analysis of WT and deletion mutant S. pombe strains. Cells were nitrogen-starved prior to analysis (24 h in EMM2-N medium) to induce EGT synthesis.