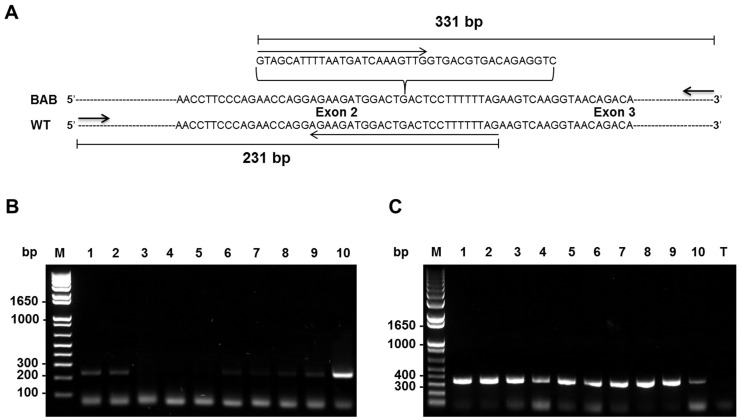
Figure 7. Residual wild-type transcripts.
(A) Partial sequence at exon2/exon3 junction of myostatin cDNA in Blonde d'Aquitaine (BAB) and normal (WT) cattle. Extra-exonisation in BAB at exon2/exon3 (G/A) junction is indicated (bracket). Two specific primers (PCRWT-F/PCRWT-R and PCRBAB-F/PCRBAB-R, Table 2) were used to amplify either the correctly spliced (B) transcript (WT strand) or (C) the transcript with an extra exon (BAB strand). (C) The aberrant transcript was detected at relatively higher levels as compared to the heterozygous G/T animal (line 10) in agreement with real-time RT-qPCR data (Fig. 6). Of note, residual correctly spliced transcripts were detected in G/G animals at variable levels ranging from undetectable (lines: 3, 4, 5) to detectable levels (lines: 1, 2, 6, 7, 8, 9) as compared to the G/T heterozygous animal (line 10). T: PCR assay without sample; M: size marker.