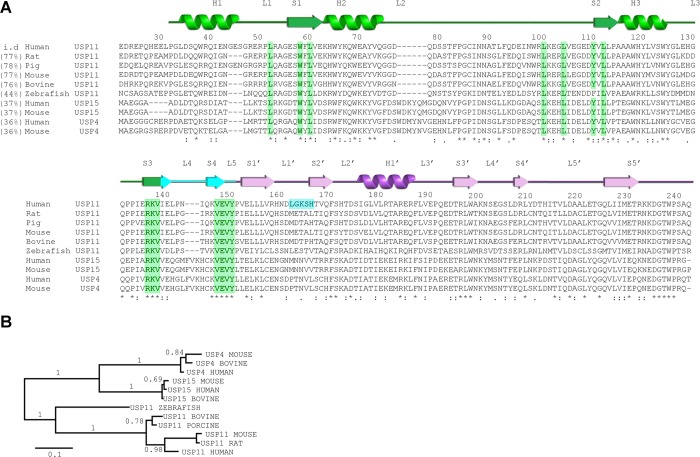
Figure 2.
Sequence conservation in USP11 and the homologues USP15 and USP4. (A) Multiple sequence alignment of the N-terminal DUSP-UBL domain region of human USP11 (UniProt P51784), porcine USP11 (UniProt F1RWV6), mouse USP11 (UniProt Q99K46), bovine USP11 (UniProt A5PKF9), rat USP11 (UniProt Q5D006), zebrafish USP11 (UniProt F1QPF4), human USP15 (UniProt Q9Y4E8), mouse USP15 (UniProt Q8R5H1), human USP4 (UniProt Q13107) and mouse USP4 (UniProt P35123) generated using Clustal Omega. Residue numbers and secondary structure elements are based on the structure of hUSP11DU assigned using the PDBsum server at the EBI. Sequence identity of respective sequences shared with human USP11 are given in brackets. α-Helices (H) and β-strands (S) from the UBL domain are renumbered to match nomenclature for ubiquitin and as such are denoted S′ or H′. Residues involved in the DUSP-linker interface described in Figure 1C are highlighted in green. The loop connecting strand S1′ and S2′ of the UBL domain (shown in blue) is not conserved in other USP11 sequences. (B) Phylogenetic analysis of members of the DU family show that the evolutionary divergence is greater among USP11 orthologues compared to USP15 and USP4 orthologues.