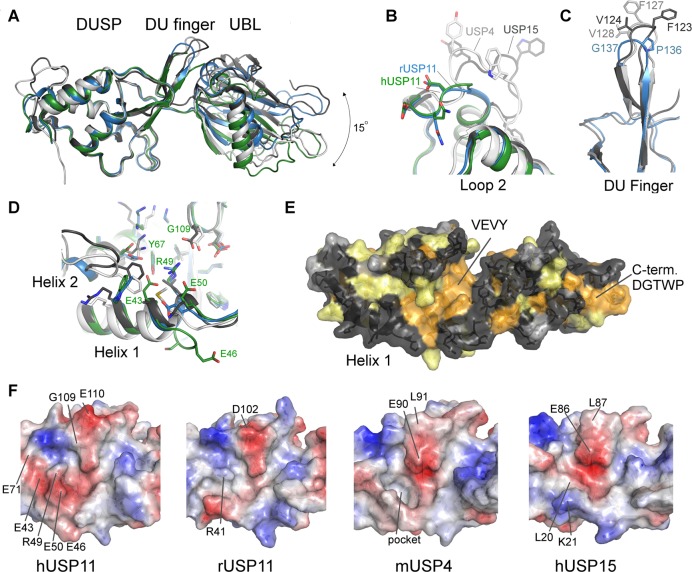
Figure 3.
Structural features of USP11, USP4, and USP15 DU domains. (A) Superposition of the structures of hUSP11DU (green), rUSP11DU (blue), hUSP15DU (dark gray, PDB code 3T9L(23)), mUSP4 (light gray, PDB code 3JYU, SGC). The structures are aligned using the DUSP domain as reference and show that the relative orientation of the UBL domain varies by approximately 15 degrees. Several important surface features are highlighted in close-up views in B, C, and D. (B) Loop 2 following helix 2 in the DUSP domain is locally the most structurally diverse region in members of the DU family. (C) The DU finger region in rUSP11 is three residues shorter than in USP15 and USP4 and is capped by a proline and glycine residue, whereas in USP15 and USP4 the fingertip harbors phenylalanine and valine residues. (D) Helix 1 in the DUSP domain is three residues longer in hUSP11 compared to rUSP11 and other members of the DU family. A GESG motif breaks the helix. Residues in helix 1 may be functionally significant as they contribute to a hydrophobic cleft on the DUSP surface in hUSP15 and mUSP4. The presence of an arginine (R49) and two glutamic acid residues (E43, E50) significantly alter the physicochemical properties of this surface feature in hUSP11DU. (E) Surface representation of the rUSP11DU structure colored according to sequence conservation in USP11, USP4, and USP15. Identical residues are colored orange, similar residues yellow, weakly similar in light gray, and dissimilar in dark gray. (F) Surface representations colored according to electrostatic potential of the DUSP domain cleft region in hUSP11, rUSP11, hUSP15, and mUSP4 with the same face of the DUSP domain shown as in (D) and (E). Residues not conserved between the structures that contribute to different surface characteristics are labeled as well as the deep hydrophobic pocket in USP4.