Figure 6. Expression of the de-acetylation mimicking mutant of CTTN/9KR in shNEDD9 cells is sufficient to rescue lamellapodia stability.
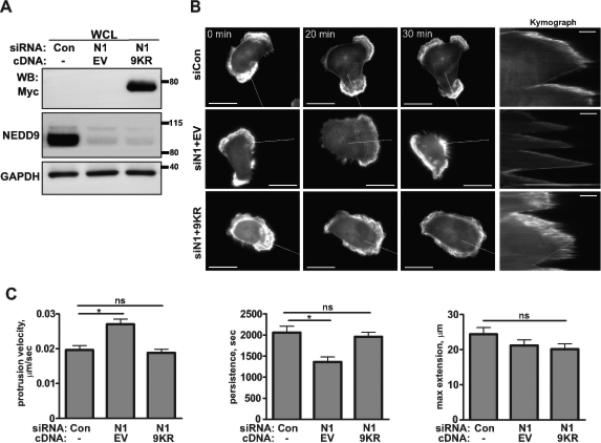
A. WB analysis of myc-CTTN/9KR mutant or empty vector (EV) expression in MDA-MB-231 cells transfected with siRNAs against NEDD9 (N1) or non-targeting control (Con) with anti-myc, -NEDD9 and -GAPDH antibodies. B. Representative images of the MDA-MB-231 cells co-trasfected with siControl (siCon); or siNEDD9 (siN1) and pRK5-myc-empty vector (siN1+EV); or siNEDD9 and pRK5-myc-CTTN/9KR (siN1+9KR) and LifeAct-mCherry (white) were used to generate composite kymographs (inserts on the right, scale bar 5μm). White lines indicate the section across the cell membrane used to quantify the velocity, persistance/stability and extension of protrusions as described in Supplemetary methods. The scale bar is 20μm. C. Quantification of protrusion velocity, persistance and maximum extension in cells as in A-B. Data presented as a mean values ± S.E.M from three independent experiments, n=10 per treatment group. One-way ANOVA with pairwise comparison for velocity: *p=0.0004 for siCon/siN1+EV, p is ns for siCon/siN1+9KR; persitance: *p=0.0003 for siCon/siN1+EV, p is ns for siCon/siN1+9KR; maximum extension: p is ns for siCon/siN1+EV or /siN1+9KR.
