Figure 2.
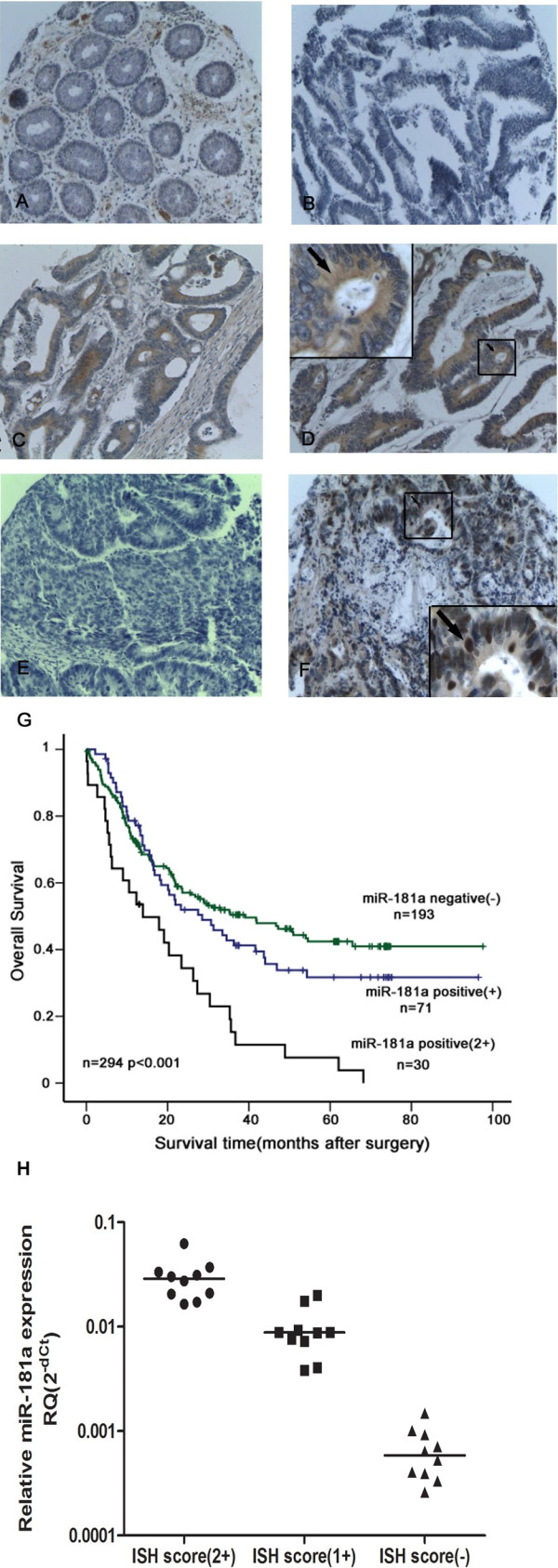
miRNA detection in colorectal cancer tissues by LNA-ISH. In situ hybridization analyses using 5′ DIG-labeled, LNA-modified DNA probes complementary to miR-181a, U6 small nuclear RNA and scrambled oligonucleotide (negative control) were performed on tissue microarray sections. (A) The expression of miR-181a was negative or weak in most of the normal colorectal mucosa; (B) Negative expression of miR-181a was also detected in some colorectal cancer tissues; (C) The weak positive expression of miR-181a was detected in the cytoplasm of cancer cells (+); (D) The strong positive expression of miR-181a was detected in the cytoplasm of cancer cells (2+); (E) No signal was detected in colorectal cancer tissues by using scrambled oligonucleotide probe (negative control); (F) The nuclear expression of U6 (positive control); (G) Association between miR-181a expression in tumors and overall survival in 294 patients with CRC using in situ hybridisation (cohort 2). Kaplan-Meier overall survival curves for CRC patients with follow-up information were compared according to miR-181a expression level. Patients with higher miR-181a expression displayed a shorter overall survival after surgery. (H) Association between miR-181a expression in CRC tumors and miR-181a ISH score. The levels of miR-181a detected by qPCR in 30 sample tissues that are matched to the samples used for ISH.
