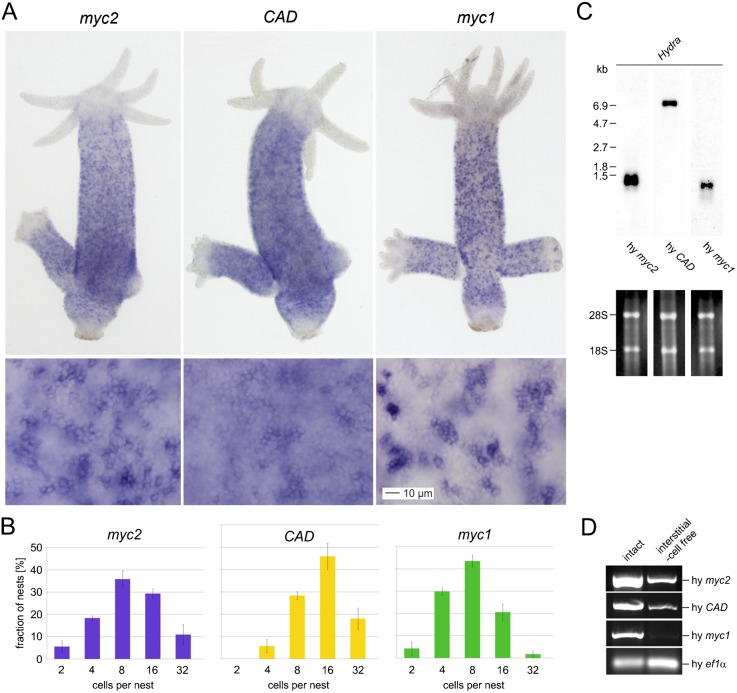
Fig. 1. Expression of the Hydra myc2, CAD, and myc1 genes.
(A) Expression patterns of myc2, CAD, and myc1 visualized by in situ hybridization. Hydra myc2, CAD, and myc1 are expressed in cells throughout the gastric region of intact, budding polyps. Higher magnifications (lower panels) from gastric regions reveal that myc2, CAD, and myc1 are up-regulated in interstitial stem cells and proliferating nematoblast nests. In addition, myc2 and CAD are expressed in the surrounding epithelial cells. (B) Quantitative analysis of myc2-, CAD-, and myc1-positive interstitial stem cells and nests of proliferating nematoblasts. The number of cells per nest was determined by visual inspection in the gastric region of in situ hybridized polyps. Each bar represents the mean ± SD from three different counts using various polyps. 100 nests were analyzed in each count. (C) Northern analyses using aliquots (2.0 µg) of poly(A)+-selected RNAs from whole Hydra animals and Hydra myc2, CAD or myc1 specific cDNA probes derived from the coding regions. Ethidium bromide-stained RNAs used for blot analysis are shown below. RNA size markers and positions of residual ribosomal RNAs (28S, 18S) are given on the left site. (D) Detection of myc2 and CAD mRNAs in reverse transcriptase-polymerase chain reaction (RT-PCR) analysis using RNA from interstitial cell-free Hydra. Amplification of a specific segment from the Hydra ef1α gene served as a control. Scale bar: 10 µm.