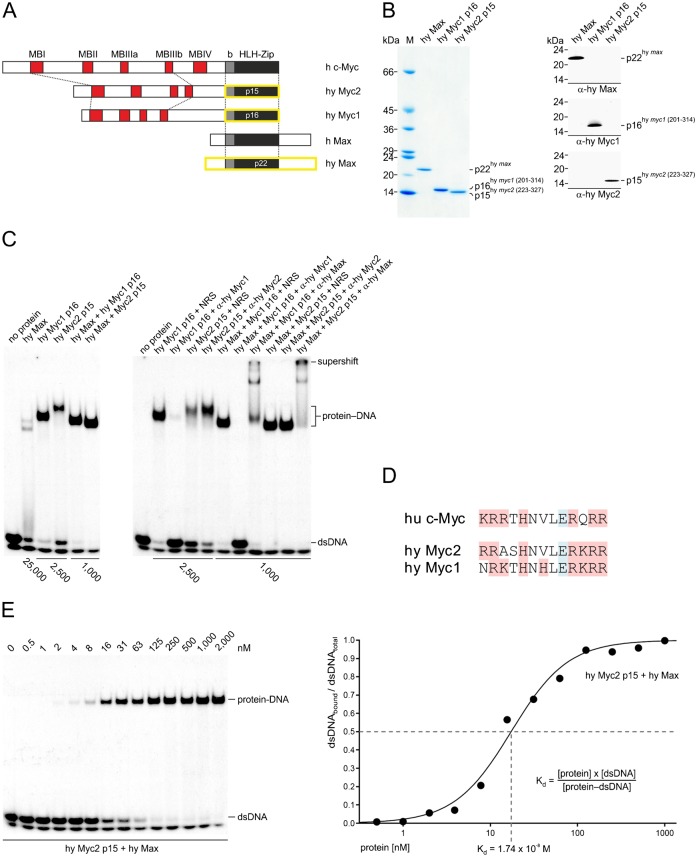
Fig. 4. Hydra Myc proteins and their biochemical activities.
(A) Schematic depiction of the human (h) c-Myc, Max, and the Hydra (hy) Myc2, Myc1, and Max protein products (GenBank accession nos.: h c-Myc, NP_002458; h Max, NP_002373; hy Myc2, ADA57607; hy Myc1, ACX32068; hy Max, ACX32069). The positions of conserved basic (b) and helix–loop–helix-zipper (HLH-Zip) domains and of the Myc boxes I–IV (MB) are indicated. The carboxyl-terminal segments of Hydra Myc2 (p15) and Myc1 (p16), and the full-length Hydra Max (p22) encompassing dimerization and DNA binding domains (b-HLH-Zip) (framed in yellow) were expressed in Escherichia coli. (B) SDS-PAGE (5.0–17.5% gradient, wt/vol) of 2-µg (Coomassie brilliant blue staining) or 50-ng (immunoblotting) aliquots of purified recombinant Hydra Myc1 p16 (amino acids 201–314), Myc2 p15 (amino acids 223–327), and Hydra Max p22. (C) Electrophoretic mobility shift assay (EMSA) using the recombinant proteins shown in panel A and 0.3-ng (25,000 cpm) aliquots of a 32P-labeled double-stranded 18-mer oligodeoxynucleotide containing the canonical Myc/Max-binding motif 5′-CACGTG-3′ in the context of an upstream stimulatory factor binding site (E-box USF). Final protein concentrations are indicated below. Left panel: DNA binding of Max, Myc1 p16 and Myc2 p15 homodimers, or Max/Myc1 p16 and Max/Myc2 p15 heterodimers. Right panel: effects of specific antibodies (α) directed against Hydra Max, Myc1 p16, or Myc2 p15 to the binding reactions. (D) Amino acid sequence alignment of human (hu) c-Myc and the Hydra (hy) Myc2 and Myc1 basic regions. Positively and negatively charged residues are shaded in red or blue, respectively. (E) Determination of the dissociation constant (Kd) of protein–DNA complexes by EMSA analysis. Titration of a double-stranded oligodeoxynucleotide as in panel C with increasing amounts of Hydra Myc2 p15 and Hydra Max. Total protein concentrations are indicated at the top (left panel). The ratios of bound DNA to total DNA were determined by phosphorimaging and plotted versus protein concentrations (right panel). Because the experimental conditions led to partial DNA strand separation, only double-stranded DNA was considered for the quantification of unbound DNA. The sigmoidal fit function f(x) = 1/{1+exp[(a−x)/b]} was used to generate the binding curve. The calculated Kd value for the protein–DNA binding reaction is indicated below.