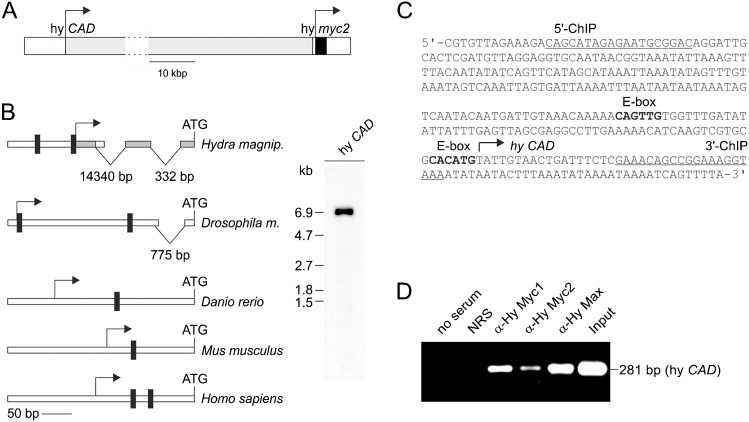
Fig. 5. Binding of Hydra Myc and Max proteins to the Hydra CAD promoter.
(A) Topography of the Hydra magnipapillata genomic locus (accession nos. NW_002159242, NW_002159544), containing the hy CAD and myc2 genes. The transcription start sites mapped by 5′RACE are indicated by arrows. (B) Left site: schematic structure of the Hydra magnipapillata CAD promotor compared with representatives from invertebrate (drosophila melanogaster) or vertebrate (danio rerio, mus musculus, homo sapiens) organisms. The positions of consensus Myc binding sites (CANNTG), of the transcription start sites (arrows), and of introns are indicated. Right site: Northern analysis using 2.0 µg of poly(A)+-selected RNA from whole Hydra animals, and the polymerase chain reaction (PCR) product (shaded in grey) obtained from 5′-rapid amplification of Hydra CAD cDNA ends as a probe. The probe specifically detects the 7.0-kb Hydra CAD mRNA. (C) Nucleotide sequence of the Hydra CAD regulatory region. The transcription start site (arrow), potential binding sites (bold) for the transcription factor Myc (CANNTG), and binding sites for 5′ and 3′ chromatin immunoprecipitation (ChIP) primers (underlined) are indicated. (D) ChIP using chromatin from whole Hydra animals. Antisera directed against Hydra Myc1, Myc2, and Max were used for precipitation, followed by PCR amplification of a 281-bp fragment from the CAD promoter. Reactions with no antiserum, normal rabbit serum (NRS), or total chromatin (Input) were used as controls.