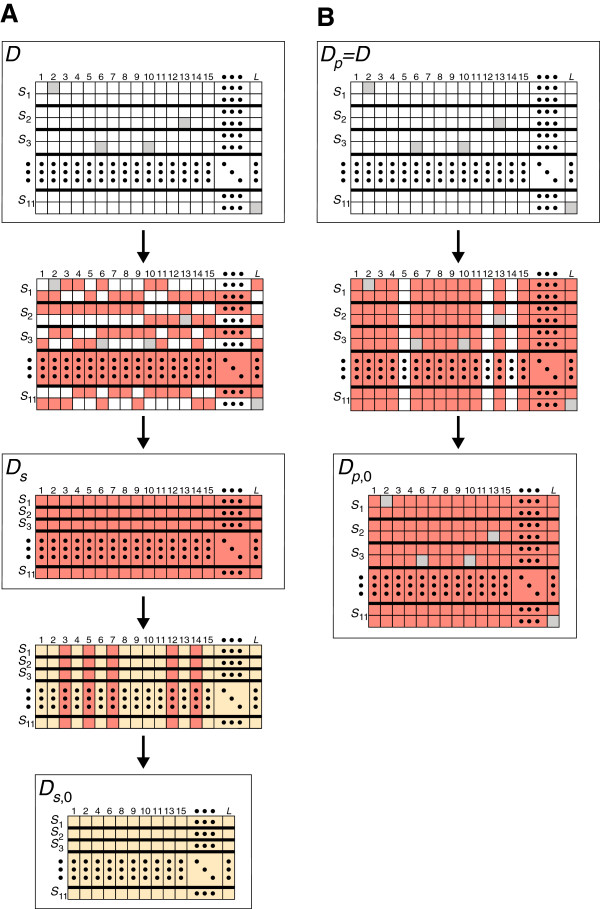
Figure 2.
Schematic for creating the four subsets,,, and from dataset . For the matrices of datasets
. For the matrices of datasets  , , , , and (see Table 2), each row is an individual and each column is a locus. Thick black lines in these matrices separate the individuals in different species. Gray boxes indicate missing sequences. (A) At each locus, a single sequence from each species (indicated in red) is selected from dataset
, , , , and (see Table 2), each row is an individual and each column is a locus. Thick black lines in these matrices separate the individuals in different species. Gray boxes indicate missing sequences. (A) At each locus, a single sequence from each species (indicated in red) is selected from dataset  . These selected sequences are used to create such that there exists a single sequence sampled per species at each locus. Sequences from a subset of loci in (indicated in yellow) are used to create dataset such that each locus has at least one nucleotide difference between each distinct pair of species other than pairs from distinct outgroups. (B) Dataset is the full starting dataset
. These selected sequences are used to create such that there exists a single sequence sampled per species at each locus. Sequences from a subset of loci in (indicated in yellow) are used to create dataset such that each locus has at least one nucleotide difference between each distinct pair of species other than pairs from distinct outgroups. (B) Dataset is the full starting dataset 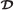 . At each locus ℓ, a distance matrix is created according to eq. 2. Sequences from a subset of loci (indicated in red) in are used to create dataset such that each locus has a nonzero p-distance between each distinct pair of species other than pairs from distinct outgroups. Observe that the matrix includes loci 3 and 7, which are not included in the matrix. Loci 3 and 7 are included in but not in because in , pairs of species contain at least one pair of individuals with different sequences, whereas in , at least one pair of the 11 selected individuals have identical sequences. Therefore, the set of loci in is a superset of the set of loci in , and the number of loci in is always greater than or equal to the number of loci in .
. At each locus ℓ, a distance matrix is created according to eq. 2. Sequences from a subset of loci (indicated in red) in are used to create dataset such that each locus has a nonzero p-distance between each distinct pair of species other than pairs from distinct outgroups. Observe that the matrix includes loci 3 and 7, which are not included in the matrix. Loci 3 and 7 are included in but not in because in , pairs of species contain at least one pair of individuals with different sequences, whereas in , at least one pair of the 11 selected individuals have identical sequences. Therefore, the set of loci in is a superset of the set of loci in , and the number of loci in is always greater than or equal to the number of loci in .