Abstract
Background:
The transcription factor, zinc finger protein 143 (ZNF143), positively regulates many cell-cycle-related genes. The ZNF143 would show high expression of multiple solid tumours related closely to cancer cell growth, similar to the widely accepted Ki67 (MIB-1) protein, but the underlying mechanisms for ZNF143 remain unclear. We investigated the association of ZNF143 expression with clinicopathological features and prognoses of patients with lung adenocarcinoma.
Methods:
Expressions of ZNF143 and MIB-1 were immunohistochemically analysed in 183 paraffin-embedded tumour samples of patients with lung adenocarcinoma. The ZNF143 expression was considered to be strong when >30% of the cancer cells demonstrated positive staining.
Results:
Strong ZNF143+ expression showed a significantly close relationship to pathologically moderate to poor differentiation and highly invasive characteristics. The ZNF143 positivity potentially induced cell growth of lung adenocarcinoma, correlated significantly with high MIB-1 labelling index (⩾10%). Univariate and multivariate analyses demonstrated that both strong ZNF143+ and the high MIB-1 index group have only and significantly worse survival rates.
Conclusions:
The combination of strong ZNF143 expression and high MIB-1 index potentially predicts high proliferating activity and poor prognosis in patients with lung adenocarcinoma, and may offer a therapeutic target against ZNF143.
Keywords: lung adenocarcinoma, ZNF143, cell growth, MIB-1, prognosis
Lung cancer is one of the most lethal malignancies in the majority of developed countries (Jemal et al, 2011; Mitsudomi et al, 2013), and it has been the most common cause of cancer-related deaths among Japanese since 1998. In fact, ∼50 000 new cases of lung cancer are diagnosed each year; 20 000 patients die of it in Japan alone (http://ganjoho.jp/professional/index.html, 2012). Adenocarcinoma is most common type of lung cancer in Japan, and in up to 30% of the patients, it recurs within 5 years even with pathological stage I after curative complete surgical resection (Asamura et al, 2008). It is conceivable that potential cell growth of lung cancer with or without occult metastases at the time of surgical intervention is primarily responsible for its poor prognosis (Yamashita et al, 2010). Therefore, predicting which patients are prone to develop recurrence and mortality after surgery is critical, as the clinical picture of lung cancer is considered to be strongly determined by the complex interplay of additional cellular alterations, for example, epigenetic modulation of gene expression (Osada and Takahashi, 2002). Actually, clinicopathological factors, such as the tumour differentiation or the tumour–node–metastasis (TNM) stage, can indicate lung adenocarcinoma prognosis (Spiro and Silvestri, 2005; Ou and Zell, 2008), whereas molecular and genetic factors have not notably done so; the clinical significance of biological markers is under evaluation.
Cancer progression in tumours is closely related to alterations particularly in G2/M cell cycle arrest and apoptosis (Torigoe et al, 2005; Kohno et al, 2010). The cell cycle is strictly regulated by various mechanisms to ensure cell division and, thus, the transcriptional regulation of cell-cycle-related genes expression requires the orchestrated recruitment of transcription factors, but it remains to be elucidated. We and others have previously revealed that a transcription factor, zinc finger protein 143 (ZNF143), which is a human homologue of the Xenopus transcriptional activator, Staf, positively regulates many DNA replication and cell-cycle-associated genes (Myslinski et al, 2007; Izumi et al, 2010; Hernández-Negrete et al, 2011). In fact, according to the DNA microarray analysis with ZNF143-specific small interfering RNA (siRNA) transfection, 41 (27.0%) genes were categorised as concerned with the cell-cycle/DNA replication, such as Aurora kinase B or High-mobility group box 2, among the <0.4-fold downregulated 152 genes (Izumi et al, 2010). For example, ZNF143 plays a central role in regulating the mitochondrial transcriptional initiation and replication of mitochondrial DNA (Gérard et al, 2007). Indeed, ZNF143 is a critical regulator of mammalian normal development and embryonic stem cell renewal (Chen et al, 2008; Chia et al, 2010; Halbig et al, 2012). In contrast, it is specifically expressed in multiple solid tumours, including lung cancer, oesophageal and gastric cancer, urothelial carcinoma, or cerebral astrocytoma (Izumi et al, 2010). Various collecting data in our group also demonstrated that cell growth of lung cancer cell lines was significantly correlated with ZNF143 expression, whereas downregulation of ZNF143 strongly induced apoptosis following G2/M cell cycle arrest (Izumi et al, 2010). It is very likely that ZNF143 might not only regulate basic cellular functions at the transcriptional level, but be the parameter of cell proliferative activity, very similar to the widely accepted Ki67 (MIB-1) protein (Burger et al, 1986; Oka et al, 2011), and subsequently be promising targets for lung cancer diagnoses and therapies. However, we have seen no studies of possible associations between ZNF143 expression in lung adenocarcinoma and its clinicopathological features, including tumour differentiation and invasiveness or patient prognoses.
In the current study, using a specific polyclonal antibody against ZNF143, we evaluated the expression of ZNF143 in lung adenocarcinoma specimens with clinicopathological data of the patients, showing that ZNF143 was strongly expressed in most human lung adenocarcinomas, especially those with poor differentiation and highly invasive characteristics, and >10% MIB-1 labelling index (i.e., high proliferating activity). Furthermore, the combination of strong ZNF143+ expression and high MIB-1 index has been only and significantly correlated with poor outcome.
Materials and methods
Patients and tissue specimens
All the intended procedures of the present study, including use of specimens from human subjects, were approved by the Ethics Committee of University of Occupational and Environmental Health (UOEH) in Kitakyushu, Japan. Pathological reports were reviewed to identify patients who underwent lobectomy (165 patients), pneumonectomy (5 patients), partial resection (4 patients), or segmentectomy (9 patients) for lung adenocarcinoma, respectively, between 1997 and December 2004 at the hospital of UOEH. Patients who suffered perioperative deaths, defined as death during the patient's initial hospitalisation or within 30 days of surgery, were excluded. A total of 183 patients with available follow-up data comprised the cohort of this retrospective study, after further excluding those with the following characteristics: (1) other prior or concomitant malignant tumours; (2) coexisting medical problems of sufficient severity to shorten life expectancy; and (3) adjuvant chemotherapies or radiotherapies before the surgery. Three pathologists examined all resected specimens to confirm their histopathological features, including differentiation. Revisions in the International System for Staging Lung Cancer was used for final staging (Vallières et al, 2009); and all lung adenocarcinomas were further classified based on the histological classification system from the International Association for the Study of Lung Cancer (IASLC)/American Thoracic Society (ATS)/European Respiratory Society (ERS) international multidisciplinary classification of lung adenocarcinoma (Travis et al, 2011).
In accordance with this IASLC/ATS/ERS classification system (Travis et al, 2011), adenocarcinoma in situ (AIS) cases were selected in haematoxylin and eosin (H&E)-stained sections, according to the following criteria: localised lesion (⩽3 cm) with growth of neoplastic cells along pre-existing alveolar structures; lack of stromal invasion; absence of papillary or micropapillary patterns; and absence of intra-alveolar tumour cells. Tumours were subclassified as minimally invasive adenocarcinoma (MIA) in cases with a solitary adenocarcinoma (⩽3 cm) with a predominantly lepidic growth pattern and ⩽5 mm invasion in the greatest dimension of any one focus. The invasive component to be measured in MIA was defined as follows: histological subtypes other than a lepidic pattern (i.e., acinar, papillary, micropapillary, or solid) or tumour cells infiltrating myofibroblastic stroma. The invasive component was measured morphometrically and a 5 mm cutoff was used to distinguish MIA from lepidic predominant invasive adenocarcinoma (LPA). For cases that contained multiple tumour foci, only the largest tumour focus was examined. Elastica van Gieson (EVG) stains were also performed if necessary. The MIA was excluded if the tumour invaded the lymphatics, blood vessels, pleura, or contained tumour necrosis. The LPA and nonlepidic adenocarcinomas with >5 mm invasion in diameter were classified as invasive adenocarcinoma and were divided further into acinar (APA), papillary (PPA), solid (SPA), and micropapillary (MPA) based on their predominant invasive pattern in H&E sections.
Clinical information was gathered from the records of patients. Survival duration was based on the date of surgery until patient's death or most recent clinic visit. Patients were followed-up and prospectively evaluated every month within the first postoperative year and at ∼2–4 months of intervals thereafter using chest X-ray, thoracic and abdominal CT scan, brain MRI, serum biochemistry, or measurements of tumour markers. The CT, MRI, and bone scintigraphy were performed every 6 months for 3 years after surgery. Additional examinations were performed if any symptoms or signs of recurrence were recognised. Formalin-fixed, paraffin-embedded tissue blocks came from our Department of Pathology. Normal human tissue was taken from nontumour portions of surgically resected specimens, and then stained with H&E, EVG, or immunohistochemistry preparations in sequential sections. The EVG and immunohistochemical D2-40 (Nichirei Bioscience Co., Tokyo, Japan, diluted 1 : 1) staining very clearly revealed pleural involvement (pl) and vascular invasion (v) in the former, and lymphatic invasion (ly) in the latter, respectively.
Preparation of antibody against ZNF143
Polyclonal antibody was raised against ZNF143 by multiple immunisations of New Zealand white rabbits with a synthetic peptide, based on the previously published work (synthetic peptide sequences: MLLAQINRDSQGMTEFPGGGMEAQHVTLC and QLGEQPSLEEAIRIASRIQQGETPGLDD; Izumi et al, 2010). The specificity of our antibody was confirmed by western blotting, demonstrating that ZNF143-specific antibody identified both endogenous 90 kDa protein and exogenous 3xFlag-ZNF143 protein, but endogenous ZNF143 protein also was completely deleted with treatment of specific ZNF143 siRNA (Izumi et al, 2010). For immunohistochemistry of ZNF143, we used human cancer cells of well to moderately differentiated squamous cell carcinoma of the lung as positive controls (Izumi et al, 2010).
Immunohistochemistry of tissue samples
Immunohistochemical staining was performed by the antibody-linked dextran polymer method for antibody-bridge labelling, with haematoxylin counterstaining (EnVision; DAKO Cytomation Co., Glostrup, Denmark). Deparaffinised and rehydrated 4-μm sections were incubated in 10% H2O2 for 5 min to block endogenous peroxidase activity. The sections were thereafter rinsed and incubated with rabbit polyclonal anti-ZNF143 (diluted 1 : 200) and mouse monoclonal MIB-1 (Ki67; Dako, diluted 1 : 50) antibodies for 30 min, respectively. The second antibody-peroxidase-linked polymers were then applied, and the sections were incubated with a solution consisting of 20 mg of 3.3′-diaminobenzidine tetrahydrochloride, 65 mg of sodium azide, and 20 ml of 30% H2O2 in 100 ml of Tris-HCL (50 mM, pH 7.6). After counterstaining with Meyer's haematoxylin, sections were observed under a light microscope. The sections were first scanned at low power for all the fields (original magnification: × 40) with tumour and nontumour tissues, respectively, to account for the heterogeneity of distribution. The number of cells showing positive nuclear staining and the pattern of staining were recorded. Necrotic tissues, stromal cells, and lymphoid cells were not included in the recording. For ZNF143 immunohistochemistry of lung adenocarcinoma specimens, positive areas comprising <9% of the neoplasms were considered negatively stained. For nuclear ZNF143 expression, positive areas that were ⩽10% were considered positively stained and were graded into three categories: weak, positive area of 10–29% strong, 30–79% and very strong, >80% positive areas. Finally, as to ZNF143 expression, all patients were divided into two groups as follows: strong when >30% whereas weak when <29%. We additionally performed immunohistochemistry on the same surgical sections in human lung adenocarcinoma tissue specimens, using newly applied and commercially available anti-ZNF143 mouse monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1 : 50 diluted). As the staining patterns and expression profiles of this monoclonal anti-ZNF143 antibody (data not shown) were very similar to those of our original polyclonal one, we can confirm that the present anti-ZNF143 antibody provided here is alternatively applied to the ZNF143 immunohistochemical examination.
The distribution of the staining with MIB-1 in the tumour and the adjacent nonneoplastic bronchioloalveolar epithelium in each case was assessed semiquantitatively and compared (Oka et al, 2011). The MIB-1 staining was divided into categories according to the percentage of the cells stained (0–4%, 5–9%, 10–29%, and 30–100%). The sections stained with MIB-1 were then counted at high power (original: × 400) magnification. At least 1000 nuclei were counted in each section. The MIB-1 labelling index was presented as number of positive nuclei per 1000 nuclei counted (Oka et al, 2011). Also, as to MIB-1 expression, all patients were divided into two groups as follows: high when >10% labelling index whereas low when <9%.
We selected and validated immunohistochemical cutoff scores for ZNF143 positivity (30%) and MIB-1 labelling index (10%) based on the performance of receiver operating characteristic (ROC) curve analysis, respectively (Hanley, 1989).
All histological and immunohistochemical slides were evaluated by two independent observers (certified surgical pathologists in our department) using a blind protocol design (observers were blinded to the clinicopathological data). Agreement between observers was excellent (>0.9) for all antibodies investigated as measured by interclass correlation coefficient. For the few instances of disagreements, a consensus score was determined by the third board-certified pathologists in our department (Li et al, 2011; Wu et al, 2012; Kitada et al, 2013).
Statistical analysis
Significance of correlations was calculated by χ2-test to assess relationships between immunohistochemical expressions and clinicopathological variables. Survival curves were plotted with the Kaplan–Meier method and compared with the log-rank test. Hazard ratios and 95% confidence intervals (95% CIs) were estimated using univariate or multivariate Cox proportional hazard models. All statistical tests were two tailed with P<0.05 considered significant. All above statistical analyses were performed with EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), the graphical user interface for R (The R Foundation for Statistical Computing, version 2.13.0, Vienna, Austria) (Kanda, 2013; Kitada et al, 2013). More precisely, it is a modified version of R commander (version 1.6-3) designed to add statistical functions frequently used in biostatistics.
Results
Patient characteristics
The cohort included 183 patients (99 men and 84 women) with clinicopathological features representative of lung adenocarcinoma (Table 1). Average age at surgery was 65 years. More than half of patients (101 out of 183) were under 400 in Brinkman index (BI); the remaining patients (82 out of 183) were ⩾400 BI. Median tumour size was 2.7 cm (range: 0.5–13.5 cm). At diagnosis, 47 (25.7%) patients had lymph node metastases and 3 (1.6%) had distant metastases. The tumour grading included 70 well differentiated (38.3%), 85 moderately differentiated (46.4%), and 28 poorly differentiated adenocarcinomas (15.3%). By further histopathological analyses with the IASLC/ATS/ERS classification system (Travis et al, 2011), 39 (21.3%) patients were AIS, 19 (10.4%) were MIA, 24 (13.1%) were LPA, 26 (14.2%) were APA, 43 (23.5%) were PPA, 3 (1.6%) were MPA, and 29 (15.8%) were SPA. Based on the international TNM classification system (seventh edition of the Union for International Cancer Control (UICC)) (Vallières et al, 2009), 124 patients had stage I disease. Postoperative follow-up was available for all 183 patients (average: 66.0 months; range: 1.7–167.5 months). Median disease-specific postoperative survival (DSS) duration was 66.8 months with 1- and 5-year survival rates of 95% and 85%, respectively.
Table 1. Detailed patients' clinicopathological characteristics.
| Characteristic | Patients (n=183) |
|---|---|
|
Age-yrs | |
| Average | 65 |
| Median | 66 |
| Range | 40–86 |
| <60 yrs | 122 |
| ≧60 yrs |
61 |
|
Sex | |
| Male | 99 |
| Female |
84 |
|
Brinkman index (BI) | |
| ≧400 | 82 |
| <400 |
101 |
|
Months after surgery | |
| Average | 66 |
| Median | 67 |
| Range |
2–168 |
|
Differentiation | |
| Well | 70 |
| Moderately | 85 |
| Poorly |
28 |
|
Histopathological subtype | |
| AIS | 39 |
| MIA |
19 |
|
Invasive adenocarcinoma | |
| LPA | 24 |
| APA | 26 |
| PPA | 43 |
| MPA | 3 |
| SPA |
29 |
|
Tumour size (cm) | |
| Average | 2.7 |
| Median | 2.4 |
| Range |
0.5–13.5 |
|
Tumour stage | |
| I | 124 |
| II | 13 |
| III | 43 |
| IV |
3 |
|
Regional lymph node metastasis | |
| N0 | 136 |
| N+ |
47 |
|
Distant metastasis | |
| M0 | 180 |
| M+ | 3 |
Abbreviations: AIS=adenocarcinoma in situ; APA=invasive adenocarcinoma, acinar predominant; LPA=invasive adenocarcinoma, lepidic predominant; MIA=minimally invasive adenocarcinoma; MPA=invasive adenocarcinoma, micropapillary predominant; PPA=invasive adenocarcinoma, papillary predominant; SPA=invasive adenocarcinoma, solid predominant.
ZNF143 expression in normal lung tissues and lung adenocarcinoma specimens
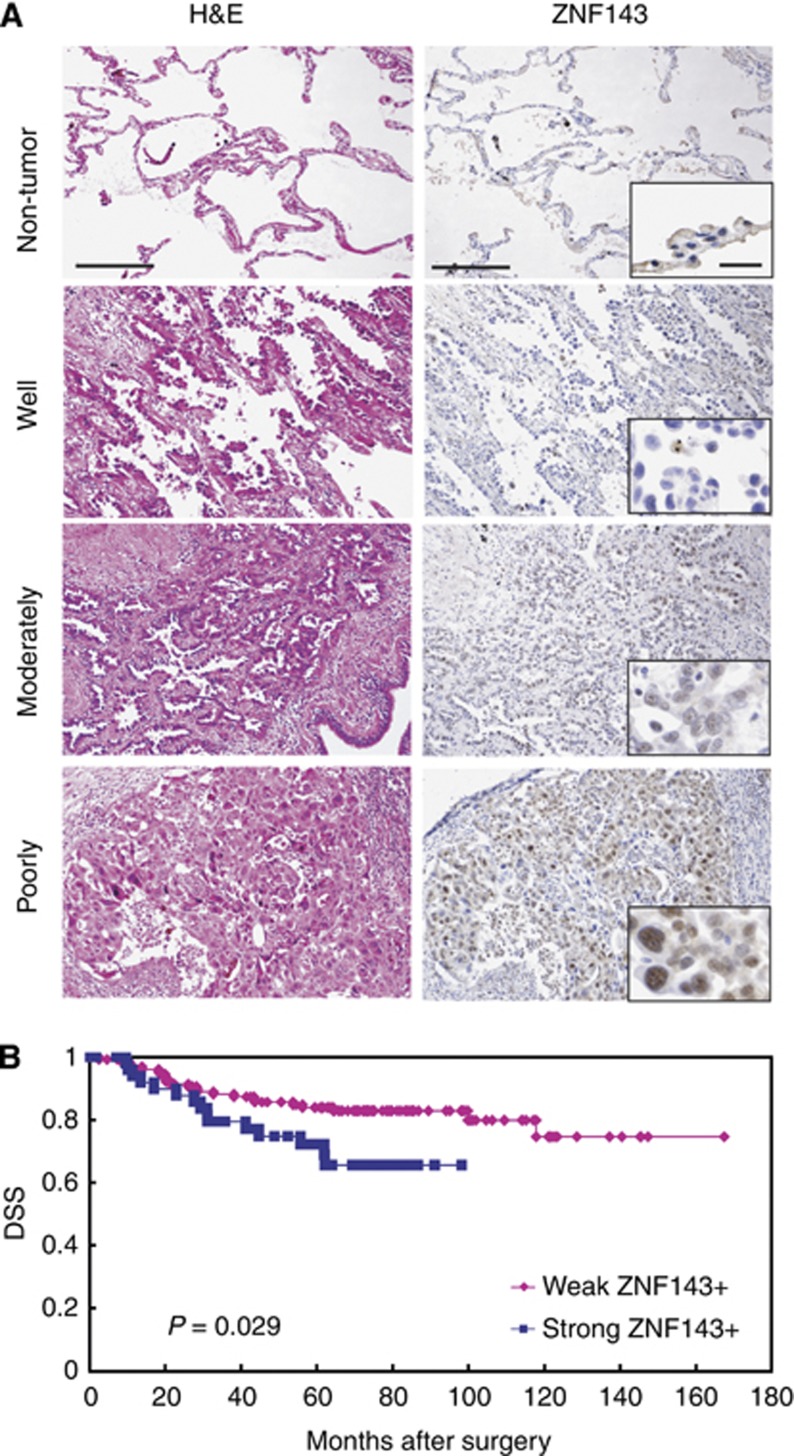
Specificity of ZNF143 polyclonal antibodies was tested using immunohistochemistry and western blotting (Izumi et al, 2010). The ZNF143 showed only nuclear immunohistochemical expression (Figure 1A). The expressions of ZNF143 were not detectable in adjacent normal bronchioloalveolar epithelium on paraffin-embedded tissues (Figure 1). The ZNF143 was weakly or strongly expressed in 131 (71.6%) or 52 (28.4%) of 183 lung adenocarcinoma specimens, respectively: 41 negative (22.4%); 90 weak (49.2%); 41 strong (22.4%); and 11 very strong (6.0%) expressions.
Figure 1.
(A) Representative images of immunohistochemical analyses of ZNF143 in human lung adenocarcinoma (well-differentiated adenocarcinoma, negative to weakly positive; moderately to poorly differentiated adenocarcinoma: strongly positive in a nuclear staining pattern) and normal bronchioloalveolar specimens (negative) (original magnification: × 100; inset, × 400). Bar=100 μm ( × 100) or 20 μm ( × 400). H&E=haematoxylin and eosin. (B) Kaplan–Meier curves of disease-specific survival (DSS) in patients with lung adenocarcinomas after surgery according to ZNF143.
Association of ZNF143 expression with clinicopathological variables
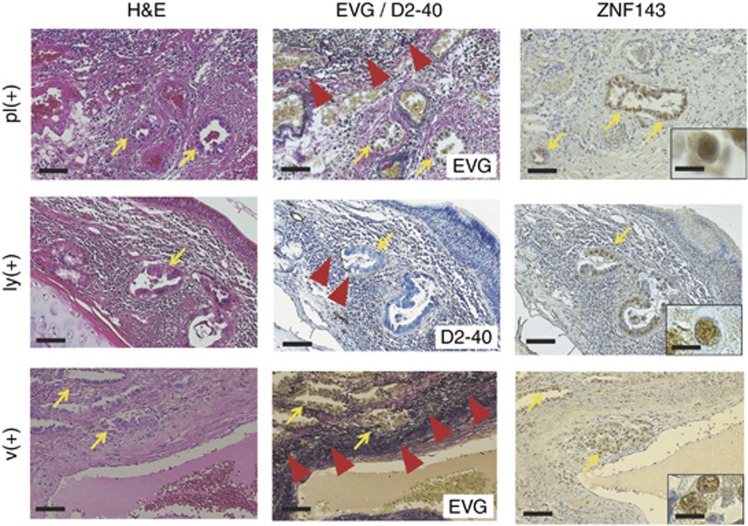
To identify the association of ZNF143 expression (weak vs strong ZNF143+) with clinicopathological characteristics of the cohort, the variables were split as shown in Table 2. There were no significant differences between patients with weak and strong ZNF143+ tumour expressions in terms of age, gender, and BI (P>0.05). However, strong ZNF143+ expression was closely associated with moderate to poor differentiation (P<0.0001), highly invasive subtypes (APA/PPA/MPA/SPA; P=0.0003), and high (⩾10%) MIB-1 labelling indexes (P=0.001; Table 2, Figure 1A, and Supplementary Figure I), but not with advanced disease stage manifesting as larger tumour size and presence of v and ly (P>0.05) in all tumours. Moreover, ZNF143 stain status borderline insignificantly correlated with the presence of pl (P=0.07). In contrast, particularly, strong ZNF143+ expression was evident in ly and v and pl of high-grade adenocarcinoma components, as shown by D2-40 and EVG stains, respectively (Figure 2). The moderately to poorly differentiated tumour rate in strong ZNF143+ samples was 39 out of 52 (75.0%), but 74 out of 131 (56.5%) in weak ZNF143+ samples (Table 2). Moreover, the highly invasive (APA/PPA/MPA/SPA) adenocarcinoma rate in strong ZNF143+ samples was 39 out of 52 (75.0%) as well, but 62 out of 131 (47.3%) in weak ZNF143+ samples (Table 2).
Table 2. Detailed correlations between ZNF143 strong expression and clinicopathological variables.
| Variables | Total, n (%) n=183 | ZNF143 strong expression, n (%) n=52 | P-value |
|---|---|---|---|
|
Age | |||
| >60 Years | 122 (66.7) | 37 (71.2) | 0.42 |
| ≦60 Years |
61 (33.3) |
15 (28.8) |
|
|
Sex | |||
| Male | 99 (54.1) | 32 (61.5) | 0.20 |
| Female |
84 (45.9) |
20 (38.5) |
|
|
BI | |||
| ≧400 | 82 (44.8) | 24 (46.2) | |
| <400 |
101 (55.2) |
28 (53.8) |
0.82 |
|
Differentiation | |||
| Well | 70 (38.3) | 13 (25.0) | |
| Moderately | 85 (46.4) | 11 (21.2) | <0.0001 |
| Poorly |
28 (15.3) |
28 (53.8) |
|
|
Histopathological subtype | |||
| AIS | 39 (21.3) | 3 (5.8) | |
| MIA | 19 (10.4) | 5 (9.6) | |
| LPA | 24 (13.1) | 5 (9.6) | |
| APA | 26 (14.2) | 5 (9.6) | 0.0003 |
| PPA | 43 (23.5) | 16 (30.8) | |
| MPA | 3 (1.6) | 1 (1.9) | |
| SPA |
29 (15.9) |
17 (32.7) |
|
|
Tumour size | |||
| >2 cm | 105 (57.4) | 32 (61.5) | 0.47 |
| ≦2 cm |
78 (42.6) |
20 (38.5) |
|
|
pl | |||
| pl(+) | 40 (21.9) | 16 (30.8) | 0.07 |
| pl(−) |
143 (78.1) |
36 (69.2) |
|
|
ly | |||
| ly(+) | 74 (40.4) | 25 (48.0) | 0.18 |
| ly(−) |
109 (59.6) |
27 (52.0) |
|
|
v | |||
| v(+) | 22 (12.0) | 7 (13.4) | 0.70 |
| v(−) |
161 (88.0) |
45 (86.6) |
|
|
pN | |||
| pN(+) | 47 (25.7) | 11 (21.2) | 0.38 |
| pN(−) |
136 (74.3) |
41 (78.8) |
|
|
pM | |||
| pM(+) | 3 (1.7) | 1 (1.9) | 0.85 |
| pM(−) |
180 (98.3) |
51 (98.1) |
|
|
MIB-1 index | |||
| ≧10% | 59 (32.2) | 26 (50.0) | 0.001 |
| <10% | 124 (67.8) | 26 (50.0) | |
Abbreviations: AIS=adenocarcinoma in situ; APA=invasive adenocarcinoma, acinar predominant; BI=Brinkman index; LPA=invasive adenocarcinoma, lepidic predominant; ly=lymphatic invasion; MIA=minimally invasive adenocarcinoma; MPA=invasive adenocarcinoma, micropapillary predominant; pl=pleural involvement; pM=distant metastasis; pN=lymph node; PPA=invasive adenocarcinoma, papillary predominant; SPA=invasive adenocarcinoma, solid predominant; v=vascular invasion; ZNF143=zinc finger protein 143. The bold and italic entries indicate significant differences (P<0.05).
Figure 2.
Representative pictures for H&E, elastica van Gieson (EVG), and immunohistochemical analyses of ZNF143and D2-40 in pleural involvement (pl) and lymphatic (ly) or vascular (v) invasion of moderately to poorly differentiated lung adenocarcinoma components (arrows; strong ZNF143+) (original magnification: × 100; inset: × 400). The EVG and D2-40 stainings very clearly reveal elastic fibres of the visceral pleura (pl(+)) or of the arterial medial wall (v(+)), and lymphatic endothelium (ly(+)), respectively (arrowheads). Bar=100 μm ( × 100) or 20 μm ( × 400).
In a Kaplan–Meier analysis (Figure 1B), lung adenocarcinoma patients with strong ZNF143+ expression had significantly shorter postoperative median DSS (58.7 months) compared with those who had weak ZNF143+ expression (72.2 months; P=0.03, Figure 1B). For patients with ZNF143+ adenocarcinomas, increasing levels of ZNF143 expression were prone to be but not significantly associated with worse clinicopathological features or longer survival (data not shown).
Association of ZNF143 expression in metastatic lung adenocarcinoma specimens of lymph nodes with prognosis
The ZNF143 was weakly or strongly expressed in 26 (55.3%) or 21 (44.7%) of 47 metastatic lung adenocarcinoma specimens of lymph nodes, respectively (Supplementary Figure 2A): 9 negative (19.1%); 17 weak (36.2%); 16 strong (34.1%); and 5 very strong (10.6%) expressions. In a Kaplan–Meier analysis (Supplementary Figure 2B), metastatic lung adenocarcinoma patients with strong ZNF143+ expression had borderline insignificantly shorter postoperative median DSS (38.1 months) compared with those who had weak ZNF143+ expression (50.4 months; P=0.08, Supplementary Figure 2B).
Association of MIB-1 expression with clinicopathological variables
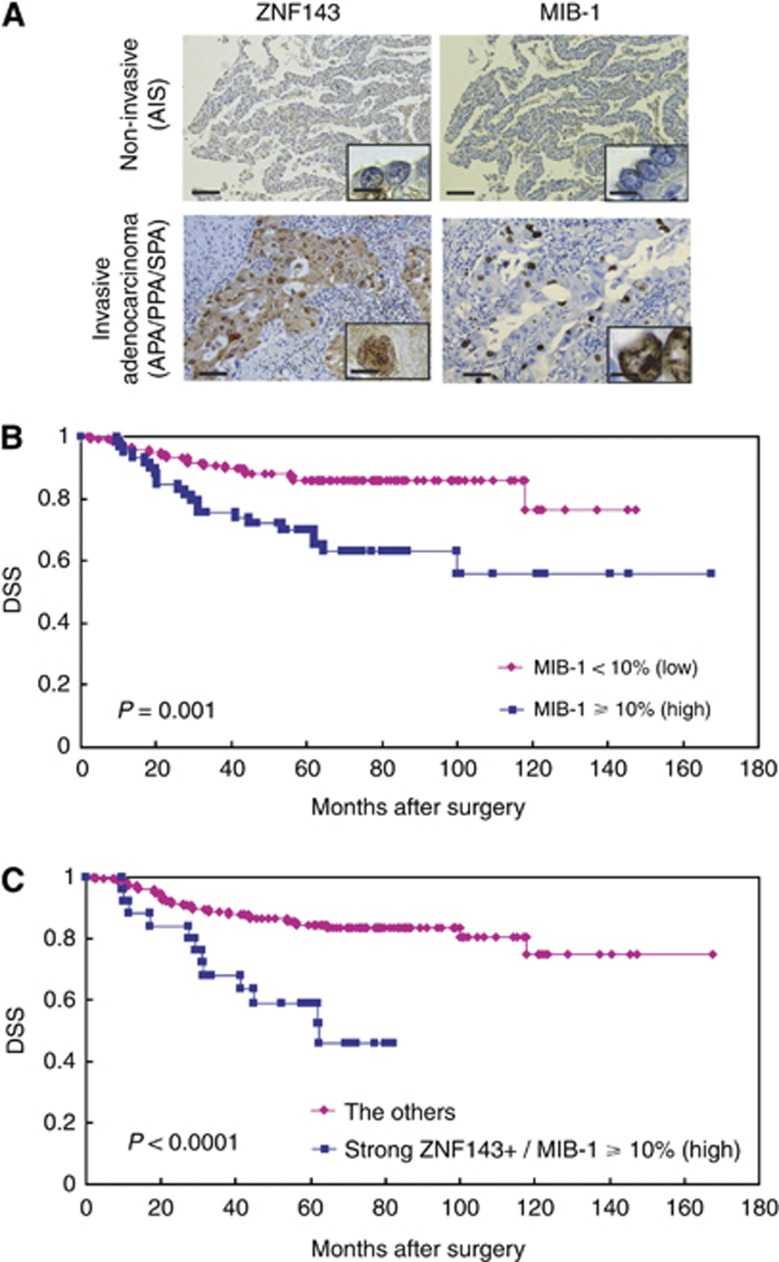
The MIB-1 also showed only nuclear immunohistochemical expression (Figure 3A). There was a heterogeneity of MIB-1 staining pattern in the lung adenocarcinomas. In well-differentiated and noninvasive or weakly invasive (AIS/MIA/LPA) adenocarcinomas, consistently, MIB-1 labelling indexes were low (<10% Figure 3A), and positive MIB-1 staining was identified only in a very small number of the relatively less differentiated cells. In contrast, its staining was significantly more diffuse and much higher in moderately to poorly differentiated or highly invasive (APA/PPA/MPA/SPA) adenocarcinomas especially in the periphery of the tumour cell nests (Figure 4A), resulting in high MIB-1 labelling indexes (⩾10%). The central carcinoma areas and adjacent tumour cells were weakly positive or negative for MIB-1, resulting in low MIB-1 indexes (<10%). Moreover, MIB-1 expressions were not detectable in adjacent normal bronchioloalveolar epithelium on paraffin-embedded tissues. The MIB-1 labelling index was >10% or <9% in 59 (32.2%) or 124 (67.8%) of 183 lung adenocarcinoma specimens, respectively: 84 (45.9%) 0–4% index; 40 (21.9%) 5–9% index; 54 (29.5%) 10–29% index; and 5 (2.7%) 30–100% index. The MIB-1 score was not related to the age or sex of the patients, BI, or tumour size, but the score was strongly correlated with the tumour grade, pl(+), ly(+), v(+), or the metastases (data not shown). Besides, in a Kaplan–Meier analysis, lung adenocarcinoma patients with high MIB-1 labelling index (⩾10%) had significantly shorter postoperative median DSS (61.9 months) compared with those who had low MIB-1 labelling index (<10%, 71.6 months, P=0.001, Figure 3B). Furthermore, increasing levels of MIB-1 labelling index score were significantly associated with worse clinicopathological features or shorter survival (data not shown).
Figure 3.
(A) Representative images of immunohistochemical analyses between ZNF143 and MIB-1 in human lung adenocarcinoma (ZNF143: noninvasive adenocarcinoma, negative to weakly positive; and invasive adenocarcinoma: strongly positive) (MIB-1 labelling index: noninvasive adenocarcinoma, low; and invasive adenocarcinoma, high) (original magnification: × 100; inset, × 400). AIS=adenocarcinoma in situ; APA=invasive adenocarcinoma, acinar predominant; PPA=invasive adenocarcinoma, papillary predominant; SPA=invasive adenocarcinoma, solid predominant. Bar=100 μm ( × 100) or 20 μm ( × 400). (B and C) Kaplan–Meier curves of DSS in patients with lung adenocarcinomas after surgery according to MIB-1 index (B) and the combination of ZNF143 and MIB-1.
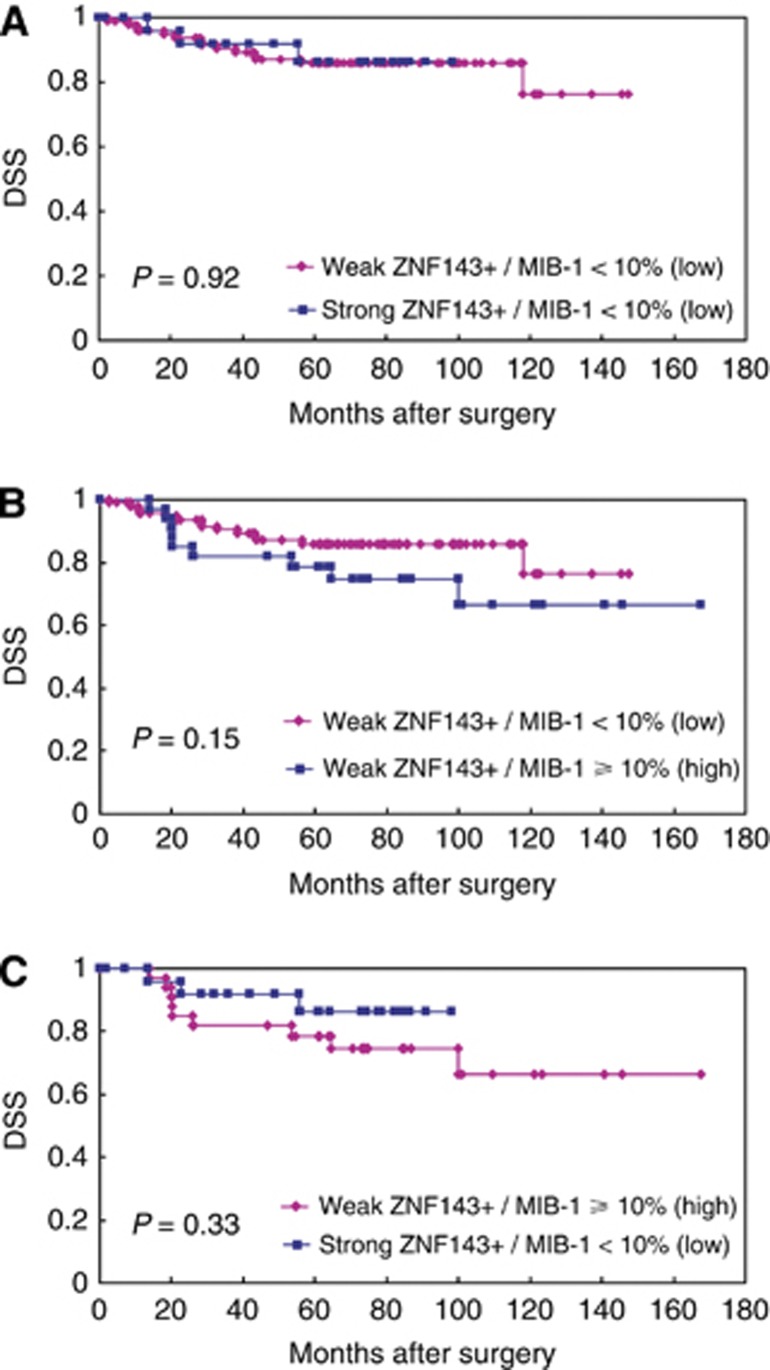
Figure 4.
Kaplan–Meier curves of DSS in patients with lung adenocarcinomas after surgery, when ZNF143/MIB-1 expressions were split into groups of either strong or weak ZNF143 expression and high or low MIB-1 labelling index, and divided into two as follows: (1) weak/low vs strong/low (A) or weak/high (B); and (2) strong/weak vs weak/high (C).
Correlations between strong ZNF143 expression and high MIB-1 labelling index
When ZNF143/MIB-1 expressions were split into groups of either strong/weak or high/low, their immunoprofiles were 53.6% weak/low (98 cases); 14.2% strong/low (26 cases); 18.0% weak/high (33 cases); and 14.2% strong/high (26 cases). First, the DSS of lung adenocarcinoma patients was divided into two: (1) weak/low vs strong/low (Figure 4A) or weak/high (Figure 4B); and (2) strong/weak vs weak/high (Figure 4C); the Kaplan–Meier method was used to verify them. The DSS of lung adenocarcinoma patients showed no significant differences (Figure 4A: P=0.92; Figure 4B: P=0.15, respectively) in the former (1) classification approach or the latter (2) (P=0.33; Figure 4C), indicating that there were neither complementary nor competitive correlations between ZNF143 and MIB-1. Next, in contrast, lung adenocarcinoma patients with strong/high expression (strong ZNF143+ and high MIB-1 index) had markedly shorter postoperative median DSS (48.4 months) compared with that of the other groups (72.1 months, P<0.0001, Figure 3C).
The combination of strong ZNF143 expression and high MIB-1 labelling index represents a significant independent prognostic indicator for lung adenocarcinoma
To assess whether ZNF143 expression was an independent predictor of postoperative DSS, a Cox proportional hazards model was created in a forward manner including only covariates that had statistically significant correlations with DSS, using an inclusion threshold of P<0.05 (Table 3). Univariate analysis showed that tumour size (>2 cm), tumour grade, presence of pl, ly, and v, lymph node (pN(+)) and distant (pM(+)) metastases, and both strong ZNF143+ and high MIB-1 labelling index status were significant predictors of poorer survival (P=0.006,<0.001,<0.001,<0.001,=0.003,<0.001,<0.001, and<0.001, respectively). Furthermore, multivariate analysis demonstrated that, after correction for confounding variables, the combination of strong ZNF143+ expression and high MIB-1 index remained an independent prognostic indicator for DSS (P=0.05) as well as distant metastasis (P=0.001).
Table 3. Univariate and multivariate analyses of survival in 183 patients with lung adenocarcinoma, according to clinicopathological variables and both strong ZNF143 expression and high MIB-1 labelling index.
| |
Univariate |
Multivariate |
||||
|---|---|---|---|---|---|---|
| Risk factors | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
| Strong ZNF143 +/MIB-1 ≧10% (high) |
3.53 |
1.77–7.04 |
<0.001 |
2.07 |
0.99–4.32 |
0.050 |
| Tumour size (>2 cm) |
2.80 |
1.33–5.91 |
0.006 |
1.27 |
0.57–2.82 |
0.56 |
| Differentiation |
6.35 |
2.25–17.87 |
<0.001 |
1.89 |
0.57–6.28 |
0.30 |
| pl(+) |
3.07 |
1.61–5.84 |
<0.001 |
1.74 |
0.89–3.40 |
0.11 |
| ly(+) |
7.23 |
3.32–15.75 |
<0.001 |
2.53 |
0.85–7.55 |
0.10 |
| v(+) |
3.03 |
1.48–6.24 |
0.003 |
1.03 |
0.45–2.36 |
0.95 |
| pN(+) |
5.52 |
2.91–10.46 |
<0.001 |
1.80 |
0.74–4.34 |
0.20 |
| pM(+) | 23.62 | 6.55–85.19 | <0.001 | 9.55 | 2.41–37.89 | 0.001 |
Abbreviations: CI=confidence interval; ly=lymphatic invasion; pl=pleural involvement; pM=distant metastasis; pN=lymph node; v=vascular invasion; ZNF143=zinc finger protein 143.
Discussion
In the current large cohort, we showed how ZNF143 expression could be closely correlated with various clinicopathological characteristics of the 183 patients with lung adenocarcinomas, using unique antihuman polyclonal ZNF143 antibody raised against the distinctive synthetic peptide. The present findings can suggest, for the first time, that a combination of ZNF143 and MIB-1 is an independent novel and powerful marker for poor prognosis in lung adenocarcinoma patients. Our collecting data reveal that strong ZNF143+ expression in lung adenocarcinoma (1) has significantly close relationship with pathological poorly differentiated characteristics or further invasive/aggressive behaviours; and (2) shows significantly high coexpression with marked MIB-1 labelling index (⩾10%), displaying potential cell growth (i.e., proliferating activity) of lung cancer. Furthermore, although our study has two limitations in its interpretation: its retrospective nature, and the fact that it was carried out at a single institution, we can present a crucial lung adenocarcinoma-specific molecule, ZNF143, that should be a poorly differentiated and invasive/proliferative tumour marker or therapeutic target for it.
Our immunohistochemical examination in lung adenocarcinoma clearly displayed a nuclear pattern for ZNF143 expression, whereas no staining was noted in adjacent normal bronchioloalveolar epithelium. This implies its potentially important roles in acquired carcinogenesis through dysregulation of DNA metabolism including cell cycle and cell growth (Izumi et al, 2010; Kohno et al, 2010). In that sense, ZNF143 should be a specific diagnostic tumour marker for lung cancer. Moreover, we have previously demonstrated that RNA interference (RNAi)-mediated downregulation of ZNF143 strongly induces apoptosis following G2/M cell cycle arrest in human prostatic adenocarcinoma (Izumi et al, 2010). On the contrary, downregulation of E2F, which is a well-known family of transcription factors to govern cell cycle/proliferation and is regulated by the binding of retinoblastoma (RB) tumour suppressor protein, induces G1 cell cycle arrest (Chen et al, 2009). Therefore, we can propose that ZNF143 comprises an ideal therapeutic target with minimal risk of side effects, as the reduction of adverse reactions caused by anticancer agents is one of the very serious issues to be resolved in clinical management. In fact, we have also reported that very weak to negative ZNF143 expressions are identified in normal nontumour tissues of the digestive tracts, urinary bladder, testis or brain (Izumi et al, 2010) as well. However, this could be a long way off, because it is difficult to predict how the various information provided here will be clinically applied. Nevertheless, much further in-depth analyses are required to clarify the utility of ZNF143 inhibitors as promising therapeutic modalities for lung adenocarcinoma.
Our obtained results are in line with in vitro studies of lung adenocarcinoma (Izumi et al, 2010) and some other carcinomas, including colon (Paek et al, 2010) and prostate (Wakasugi et al, 2007; Izumi et al, 2010) epithelial cancers, as follows: ZNF143 expression is significantly required for cell growth in human lung adenocarcinoma; and ZNF143 expression levels are conspicuously upregulated in colon or prostatic cancer cells that are resistant to cisplatin, one of the major anticancer drugs. Taken together, it is also suggested that ZNF143 should play a critical role in not only carcinogenesis but cancer cell survival, even though, to date, there have been no prior large cohort studies of ZNF143 in malignant neoplasms within our thorough investigation. Furthermore, the present study shows ZNF143 as a powerful and potentially independent negative indicator of DSS in patients with lung adenocarcinoma, especially in combination with MIB-1 index and, by extension, ZNF143 could be a novel prognostic marker for it. Reportedly, a 5-year overall survival rate is <20% in non-small-cell lung cancer (NSCLC) including adenocarcinoma (Jemal et al, 2011), and the survival rate after complete tumour resection is only 60% to 80%, even in patients with stage I NSCLC (Asamura et al, 2008; Hanagiri et al, 2010). In addition, novel targeted therapies, such as epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor or antigen-specific immunotherapy, would be of limited benefit, even after complete resection of primary lung cancer (Uramoto et al, 2010, 2013). The ZNF143 expression patterns in surgical specimens of primary but not metastatic lung adenocarcinoma might allow improved patient selection for postoperative adjuvant systemic therapies, except for cisplatin-based chemotherapy (Wakasugi et al, 2007; Kohno et al, 2010), and prediction of appropriate clinical courses after surgery. Despite that relapse within 5 years occurs in up to 30% of lung adenocarcinoma surgical cases even with pathological stage I (Asamura et al, 2008), there have been no reliable predictors of progressive potential to date. A molecular marker, ZNF143, should refine individual risk stratification and treatment plans, including the other platinum-based antineoplastic drugs or promising specific anti-ZNF143 proteins. Indeed, the current univariate and multivariate Cox survival analyses confirm that ZNF143 expression is a potentially independent prognostic factor in lung adenocarcinoma if the cutoff value is set at 30%, but not at 5% or 10%. Although there might be a possibility of data fitting in arbitrary cutoff values, the blinded protocol design and the performance of ROC curve analysis (Hanley, 1989) used here can prevent that it could have influenced the final conclusive results. Nevertheless, this discrepancy would be because of the heterogeneity of lung adenocarcinoma or the size of cohort at a single institution, at least in part, and further experiments are thus necessary to address methodology standardisation for ZNF143 in clinical specimens after collecting and investigating a much larger number of surgical cases.
In contrast, as Ki67 (MIB-1) protein is present during all active phases of the cell cycle (G1/S/G2/M) (Burger et al, 1986), we can demonstrate that the MIB-1 labelling index (⩾10%), as a well-known indicator of cancer cell growth, could also be a useful but merely adjunctive (i.e., not enough) aid for worse clinicopathological features, such as more advanced stage and shorter postoperative recurrence (Oka et al, 2011), as well as malignancy, in the lung adenocarcinomas. Actually, on one hand, MIB-1 staining properly performed would not be completely displaced as a proliferation marker, as the other marker, ZNF143, has been shown to add prognostic power to it. On the other hand, we found that strong ZNF143+ expression was closely associated with high MIB-1 positivity (P=0.001, r=0.24) in our cohort, leading to the uniquely additive effects or benefits for the combination of ZNF143 and MIB-1. It has been reported that there are at least two kinds of interaction – competition or complement – among the two representative proteins, similar to the uridine diphosphate (UDP) N-acetyl-α-D-galactosamine polypeptide (GalNAc) N-acetylgalactosaminyltransferase (GalNAc-T) family members, as proven by experiments in vitro and in vivo (Bennett et al, 1998; Brockhausen, 1999; Li et al, 2011; Kitada et al, 2013). We hypothesised that, if the relationship between strong ZNF143+ and high MIB-1 labelling index was complementary, the DSS might be different between the weak/low and strong/low or weak/high group. On the contrary, if strong ZNF143+ and high MIB-1 index competed with each other, there might be some differences in the survival duration between the strong/low group and the weak/high group. The cohort was divided in two such manners and the Kaplan–Meier method was used to verify them. Surprisingly, the DSS of patients with lung adenocarcinoma showed no significant differences in either classification approach, indicating that there were no overt competitive or complementary correlations between ZNF143 expressions and MIB-1 indexes. Based on our analyses with a large cohort, these two cancer cell growth-related proteins coexpressed but might function separately. In this context, it is very convincing that the combination of ZNF143 and MIB-1, practically used as immunohistochemical proliferative markers, only and significantly predicted clinicopathologically aggressive potential or poor outcome in patients with lung adenocarcinoma. Actually, neither ZNF143 itself nor MIB-1 itself was an independent predictive marker for survival in the present study. However, as the putative roles of these proteins in proliferation pathways seem to be very complicated and associated with expression of many other cell-cycle-related genes (Izumi et al, 2010), this study might be narrow in part for general conclusions regarding the most important synergies between them. Further follow-up cohort and molecular studies are needed to assess more comprehensive gene expression profiles and determine the intriguing relationships between ZNF143 and MIB-1.
To further elucidate ZNF143 function in lung adenocarcinoma, we examined its immunohistochemical expression patterns especially in the aggressively invasive fronts of cancer, such as vessel permeation (ly and v) or pleural involvement (pl), in which cell adhesion critically affects not only cell morphology but also cell cycle progression (Yuan et al, 2007; Hernández-Negrete et al, 2011; Kitada et al, 2013). Correspondingly, the stronger ZNF143+ expression was also prone to be found in invasive fronts of lung adenocarcinoma associated possibly with antiadhesive effects, potentially leading to poorly differentiated characteristics or invasive/aggressive behaviours, as shown here. In contrast, few recent papers have demonstrated that ZNF143 knockdown accelerates migration and invasion through an inhibitory E-cadherin-linked pathway in colon cancer cells (Paek et al, 2013), and speculated that deregulation of ZNF143 might facilitate acquisition of adhesion independence with subsequent metastases in transformed cancer cells (Hernández-Negrete et al, 2011). These conflicting data are probably because of the heterogeneity of each cancer, for example, the number of intragenetic mutations between lung and colon adenocarcinomas, in part (Vogelstein et al, 2013). Despite that, taken together, ZNF143 might have an important role in epithelial-to-mesenchymal transition (EMT) and the reverse process, mesenchymal-to-epithelial transition (MET). Much further analyses are necessary to elucidate this field; however, our original data here imply not only the key role of dysregulated cell cycle progression via strong ZNF143 expression in carcinogenesis and invasiveness/proliferation of lung adenocarcinoma, but the potential of ZNF143 as a target in lung adenocarcinoma treatment, for example, a gene therapy with RNAi for ZNF143.
In conclusion, our findings show that strong ZNF143+ expression in primary lung adenocarcinoma is very closely related to pathological phenotypes with poor prognoses, for example, those with poor differentiation and highly invasive characteristics, or high MIB-1 labelling index, manifesting as marked cancer cell growth/proliferation. Furthermore, outcomes of patients whose lung adenocarcinomas had both strong ZNF143+ and high MIB-1 index were only and significantly worse compared with those with the other groups. These analyses can indicate, for the first time, that the combination of ZNF143 and MIB-1 should be a novel and useful independent predictor of poor prognosis in patients with primary lung adenocarcinomas.
Acknowledgments
We thank Hiroko Isagai, Hana Nishimura, Naoko Une, and Mitsuhiro Kaneko for their expert technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research (24790394, 20590416, and 19590413) from the Ministry of Education, Culture, Sports, Science, and Technology, Tokyo, Japan (to SY, KK, and YS).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Asamura H, Goya T, Koshiishi Y, Sohara Y, Eguchi K, Mori M, Nakanishi Y, Tsuchiya R, Shimokata K, Inoue H, Nukiwa T, Miyaoka E, Japanese Joint Committee of Lung Cancer Registry A Japanese Lung Cancer Registry study: prognosis of 13,010 resected lung cancers. J Thorac Oncol. 2008;3 (1:46–52. doi: 10.1097/JTO.0b013e31815e8577. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Hassan H, Mandel U, Mirgorodskaya E, Roepstorff P, Burchell J, Taylor-Papadimitriou J, Hollingsworth MA, Merkx G, vanKessel AG, Eiberg H, Steffensen R, Clausen H. Cloning of a humanUDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferasethat complements other GalNAc-transferases in completeO-glycosylation of the MUC1 tandem repeat. J Biol Chem. 1998;273 (46:30472–30481. doi: 10.1074/jbc.273.46.30472. [DOI] [PubMed] [Google Scholar]
- Brockhausen I. Pathways of O-glycan biosynthesis in cancer cells. Biochim Biophys Acta. 1999;1473 (1:67–95. doi: 10.1016/s0304-4165(99)00170-1. [DOI] [PubMed] [Google Scholar]
- Burger PC, Shibata T, Kleihues P. The use of the monoclonal antibody Ki-67 in the identification of proliferating cells: application to surgical neuropathology. Am J Surg Pathol. 1986;10 (9:611–617. doi: 10.1097/00000478-198609000-00003. [DOI] [PubMed] [Google Scholar]
- Chen X, Fang F, Liou YC, Ng HH. Zfp143 regulates Nanog through modulation of Oct4 binding. Stem Cells. 2008;26 (11:2759–2767. doi: 10.1634/stemcells.2008-0398. [DOI] [PubMed] [Google Scholar]
- Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9 (11:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, Huss M, Soh BS, Kraus P, Li P, Lufkin T, Lim B, Clarke ND, Bard F, Ng HH. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468 (7321:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Gérard MA, Krol A, Carbon P. Transcription factor hStaf/ZNF143 is required for expression of the human TFAM gene. Gene. 2007;401 (1–2:145–153. doi: 10.1016/j.gene.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Halbig KM, Lekven AC, Kunkel GR. The transcriptional activator ZNF143 is essential for normal development in zebrafish. BMC Mol Biol. 2012;13:3. doi: 10.1186/1471-2199-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanagiri T, Baba T, So T, Yasuda M, Sugaya M, Ono K, So T, Uramoto H, Takenoyama M, Yasumoto K. Time trends of surgical outcome in patients with non-small cell lung cancer. J Thorac Oncol. 2010;5 (6:825–829. doi: 10.1097/JTO.0b013e3181d5e47f. [DOI] [PubMed] [Google Scholar]
- Hanley JA. Receiver operating characteristic (ROC) methodology: the state of the art. Crit Rev Diagn Imaging. 1989;29 (3:307–335. [PubMed] [Google Scholar]
- Hernández-Negrete I, Sala-Newby GB, Perl A, Kunkel GR, Newby AC, Bond M. Adhesion-dependent Skp2 transcription requires selenocysteine tRNA gene transcription-activating factor (STAF) Biochem J. 2011;436 (1:133–143. doi: 10.1042/BJ20101798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H, Wakasugi T, Shimajiri S, Tanimoto A, Sasaguri Y, Kashiwagi E, Yasuniwa Y, Akiyama M, Han B, Wu Y, Uchiumi T, Arao T, Nishio K, Yamazaki R, Kohno K. Role of ZNF143 in tumor growth through transcriptional regulation of DNA replication and cell-cycle-associated genes. Cancer Sci. 2010;101 (12:2538–2545. doi: 10.1111/j.1349-7006.2010.01725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61 (2:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kanda Y. Investigation of the freely available easy-to-use software ‘EZR' for medical statistics. Bone Marrow Transplant. 2013;48 (3:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada S, Yamada S, Kuma A, Ouchi S, Tasaki T, Nabeshima A, Noguchi H, Wang KY, Shimajiri S, Nakano R, Izumi H, Kohno K, Matsumoto T, Sasaguri Y. Polypeptide N-acetylgalactosaminyl transferase 3 independently predicts high-grade tumours and poor prognosis in patients with renal cell carcinomas. Br J Cancer. 2013;109 (2:472–481. doi: 10.1038/bjc.2013.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Uchiumi T, Niina I, Wakasugi T, Igarashi T, Momii Y, Yoshida T, Matsuo K, Miyamoto N, Izumi H. Transcription factors and drug resistance. Eur J Cancer. 2010;41 (16:2577–2586. doi: 10.1016/j.ejca.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Li Z, Yamada S, Inenaga S, Imamura T, Wu Y, Wang KY, Shimajiri S, Nakano R, Izumi H, Kohno K, Sasaguri Y. Polypeptide N-acetylgalactosaminyltransferase 6 expression in pancreatic cancer is an independent prognostic factor indicating better overall survival. Br J Cancer. 2011;104 (12:1882–1889. doi: 10.1038/bjc.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsudomi T, Suda K, Yatabe Y. Surgery for NSCLC in the era of personalized medicine. Nat Rev Clin Oncol. 2013;10 (4:235–244. doi: 10.1038/nrclinonc.2013.22. [DOI] [PubMed] [Google Scholar]
- Myslinski E, Gérard MA, Krol A, Carbon P. Transcription of the human cell cycle regulated BUB1B gene requires hStaf/ZNF143. Nucleic Acids Res. 2007;35 (10:3453–3464. doi: 10.1093/nar/gkm239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Uramoto H, Shimokawa H, Iwanami T, Tanaka F. The expression of Ki-67, but not proliferating cell nuclear antigen, predicts poor disease free survival in patients with adenocarcinoma of the lung. Anticancer Res. 2011;31 (12:4277–4282. [PubMed] [Google Scholar]
- Osada H, Takahashi T. Genetic alterations of multiple tumor suppressors and oncogenes in the carcinogenesis and progression of lung cancer. Oncogene. 2002;21 (48:7421–7434. doi: 10.1038/sj.onc.1205802. [DOI] [PubMed] [Google Scholar]
- Ou SH, Zell JA. Validation study of the proposed IASLC staging revisions of the T4 and M non-small cell lung cancer descriptors using data from 23,583 patients in the California Cancer Registry. J Thorac Oncol. 2008;3 (3:216–227. doi: 10.1097/JTO.0b013e318164545d. [DOI] [PubMed] [Google Scholar]
- Paek AR, Kim SH, Kim SS, Kim KT, You HJ. IGF-1 induces expression of zinc-finger protein 143 in colon cancer cells through phosphatidylinositide 3-kinase and reactive oxygen species. Exp Mol Med. 2010;42 (10:696–702. doi: 10.3858/emm.2010.42.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek AR, Lee CH, You HJ. A role of zinc-finger protein 143 for cancer cell migration and invasion through ZEB1 and E-cadherin in colon cancer cells. Mol Carcinog. 2013;53 (Suppl 1:E161–E168. doi: 10.1002/mc.22083. [DOI] [PubMed] [Google Scholar]
- Spiro SG, Silvestri GA. One hundred years of lung cancer. Am J Respir Crit Care Med. 2005;172 (5:523–529. doi: 10.1164/rccm.200504-531OE. [DOI] [PubMed] [Google Scholar]
- Torigoe T, Izumi H, Ishiguchi H, Yoshida Y, Tanabe M, Yoshida T, Igarashi T, Niina I, Wakasugi T, Imaizumi T, Momii Y, Kuwano M, Kohno K. Cisplatin resistance and transcription factors. Curr Med Chem Anticancer Agents. 2005;5 (1:15–27. doi: 10.2174/1568011053352587. [DOI] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, Garg K, Austin JH, Asamura H, Rusch VW, Hirsch FR, Scagliotti G, Mitsudomi T, Huber RM, Ishikawa Y, Jett J, Sanchez-Cespedes M, Sculier JP, Takahashi T, Tsuboi M, Vansteenkiste J, Wistuba I, Yang PC, Aberle D, Brambilla C, Flieder D, Franklin W, Gazdar A, Gould M, Hasleton P, Henderson D, Johnson B, Johnson D, Kerr K, Kuriyama K, Lee JS, Miller VA, Petersen I, Roggli V, Rosell R, Saijo N, Thunnissen E, Tsao M, Yankelewitz D. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6 (2:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uramoto H, So T, Nagata Y, Kuroda K, Shigematsu Y, Baba T, So T, Takenoyama M, Hanagiri T, Yasumoto K. Correlation between HLA alleles and EGFR mutation in Japanese patients with adenocarcinoma of the lung. J Thorac Oncol. 2010;5 (8:1136–1142. doi: 10.1097/JTO.0b013e3181e0b993. [DOI] [PubMed] [Google Scholar]
- Uramoto H, Yamada S, Tanaka F. Angiogenesis of lung cancer utilizes existing blood vessels rather than developing new vessels using signals from carcinogenesis. Anticancer Res. 2013;33 (5:1913–1916. [PubMed] [Google Scholar]
- Vallières E, Shepherd FA, Crowley J, Van Houtte P, Postmus PE, Carney D, Chansky K, Shaikh Z, Goldstraw P, International Association for the Study of Lung Cancer International Staging Committee and Participating Institutions The IASLC Lung Cancer Staging Project: proposals regarding the relevance of TNM in the pathologic staging of small cell lung cancer in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol. 2009;4 (9:1049–1059. doi: 10.1097/JTO.0b013e3181b27799. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339 (6127:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi T, Izumi H, Uchiumi T, Suzuki H, Arao T, Nishio K, Kohno K. ZNF143 interacts with p73 and is involved in cisplatin resistance through the transcriptional regulation of DNA repair genes. Oncogene. 2007;26 (36:5194–5203. doi: 10.1038/sj.onc.1210326. [DOI] [PubMed] [Google Scholar]
- Wu Y, Yamada S, Izumi H, Li Z, Shimajiri S, Wang KY, Liu YP, Kohno K, Sasaguri Y. Strong YB-1 expression is associated with liver metastasis progression and predicts shorter disease-free survival in advanced gastric cancer. J Surg Oncol. 2012;105 (7:724–730. doi: 10.1002/jso.23030. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Uramoto H, Onitsuka T, Ono K, Baba T, So T, So T, Takenoyama M, Hanagiri T, Oyama T, Yasumoto K. Association between lymphangiogenesis-/micrometastasis- and adhesion-related molecules in resected stage I NSCLC. Lung Cancer. 2010;70 (3:320–328. doi: 10.1016/j.lungcan.2010.02.013. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Wong S, Borrelli A, Chung MA. Down-regulation of MUC1 in cancer cells inhibits cell migration by promoting E-cadherin/catenin complex formation. Biochem Biophys Res Commun. 2007;362 (3:740–746. doi: 10.1016/j.bbrc.2007.08.074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.