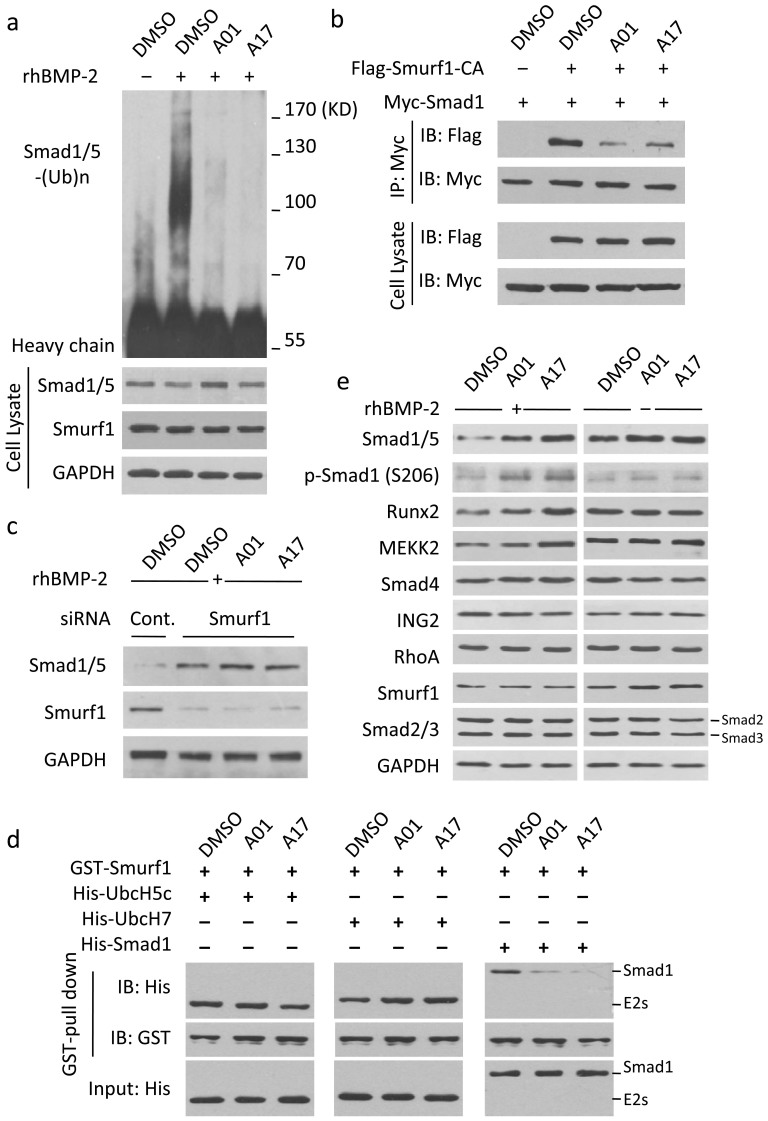
Figure 4. Selective compounds impair Smurf1-mediated Smad1/5 degradation.
(a) Selective compounds impeded the ubiquitination of Smad1/5. C2C12 cells were treated A01 and A17 at 2 μM, while MG132 and rhBMP-2 were used at 20 μM and 50 ng/ml. GAPDH were used as loading controls. Note that cropped blots are shown here. (b) Selective compounds blocked Smurf1-Smad1 interaction. 293T cells were co-transfected Flag-empty vector and Myc-Smad1 (lane 1) or Flag-Smurf1-CA and Myc-Smad1 (lane 2–4) plasmids. For inhibitors administration, cells were treated A01 and A17 at 2 μM. Note that cropped blots are shown here. (c) Selective compounds had unnoted effects on Smad1/5 without Smurf1. Smurf1 was knocked down in C2C12 cells (lane1: control siRNA, lane2–4: mouse Smurf1 siRNA). For inhibitors administration, cells were treated A01 and A17 at 2 μM, while rhBMP-2 was used at 50 ng/ml. GAPDH were used as loading controls. Note that cropped blots are shown here. (d) Selective compounds impaired Smurf1-Smad1 but not Smurf1-E2s interaction. Prokaryotic expressed proteins were purified and employed in GST-pull down. A01 and A17 were used at 10 μM. Note that cropped blots are shown here. (e) Effects of selective compounds on Smurf1 substrates and TGF-β pathway components. C2C12 cells were treated A01 and A17 at 2 μM, while rhBMP-2 was used at 50 ng/ml. GAPDH were used as loading controls. Note that cropped blots are shown here.