Abstract
The intricate relationship between angiogenesis and osteogenesis in vivo must be replicated in bone tissue engineering constructs to ensure the formation of a functional vascular network to support successful bone formation. Although communication between bone marrow stromal cells (MSC) and endothelial cells (EC) is recognized as one of the most important cellular interactions in bone regeneration, the underlying mechanisms of this biological process are not well understood. The purpose of this study was to analyze global gene expression associated with intercellular communication between MSC and EC using HumanWG-6 v3.0 expression BeadChips with a one-channel platform system (Illumina, San Diego, CA, USA). Each array contains more than 48,000 probes derived from human genes. A global map of MSC gene expression was generated following co-culture of MSC with EC for 5 and 15 days, in a direct-contact model. The map was used to determine relative alterations in functional processes and pathways. Co-culturing EC with MSC up-regulated genes related to angiogenesis as von Willebrand factor, platelet/endothelial cell adhesion molecule-1, cadherin 5, angiopoietin-related protein 4, and cell surface antigen CD34, and genes playing important roles in osteogenesis as alkaline phosphatase, FK506 binding protein 5, and bone morphogenetic protein. These findings clearly demonstrated that EC had a significant impact on MSC, particularly the bidirectional regulation of angiogenesis and osteogenesis. Moreover, cell-matrix interactions and TGF-β signal pathways were implicated for a crucial role in endothelial, cell-induced gene regulation in MSCs. A detailed study of the functional correlates of the microarray data is warranted to explore cellular and molecular interactions of importance in bone tissue engineering.
INTRODUCTION
The limited success of bone grafts has led to the development of bone constructs grown ex vivo before implantation, based upon the principles of bone tissue engineering (1). It is crucial that the engineered tissue have a fully functional vascular network at the time of implantation: this remains a major obstacle to clinical applications (2). While it is well known that limited in-growth of blood vessels from surrounding tissue into transplanted grafts will gradually occur, slow angiogenesis precludes the establishment of larger, viable grafts, since in order to receive oxygen and nutrients through diffusion, cells must be located within 100 μm to 200 μm of a blood vessel (2). Bone is formed by two distinct modes of ossification: intramembranous and endochondral (3). In intramembranous bone formation, there is an invasion of capillaries for the transport of mesenchymal stem cells (MSCs), which can differentiate directly into osteoblasts (OBs) and, in turn, secrete bone matrix. In endochondral ossification, MSCs first differentiate into chondroblasts to form a framework of cartilage, which is followed by the invasion of blood vessels that deliver specialized cells to replace the cartilage with bone and bone marrow. Another important role of vascularization in bone formation is the delivery of growth factors, which controls the recruitment, proliferation, differentiation, function, and/or survival of the bone cells. Angiogenesis precedes osteogenesis, and is an absolute requirement for osteogenesis to occur (4).
Because of the intricate association between angiogenesis and osteogenesis in vivo, communication between MSC and endothelial cells (EC) represents one of the most important cellular interactions for bone formation (2). Previous studies have shown that co-culture with endothelial cells enhances cellular proliferation of MSCs and induces differentiation into osteogenic cells with up-regulation of alkaline phosphatase (ALP) expression (5, 6). It has also been reported that EC co-cultured with OB are able to establish microcapillary-like structures in a 3D scaffold (4). Both the MSC and EC network express connexion 43 (C × 43), a specific gap junction protein, suggesting that the two cell types communicate via a gap junction channel comprising C × 43 (5). A construct comprising EC and bone marrow-derived OB, co-cultured on polycaprolactone (PCL) scaffolding, was implanted into calvarial defects in a rat model. The results revealed improved osteogenesis with enhanced vascularization (7). Applying co-cultured OB and EC within RGD-grafted alginate microspheres implanted in a long bone defect in mice showed significantly enhanced mineralization of the microspheres (2). Furthermore, bone regeneration in a rat calvarial defect was enhanced by a tissue-engineered construct of a polymer scaffold, loaded with co-cultured cells (4). The studies (4) suggest that EC can influence not only osteogenic differentiation in vitro, but also osteogenesis in vivo. Cross-talk between MSCs and ECs was identified as one of the most important cellular interactions coordinating the bone regeneration process, thus suggesting that new and multidisciplinary approaches are needed for more detailed investigations of these cellular interactions.
The aim of the present study was to analyze patterns of global gene expression associated with cross-talk between human MSCs and ECs. The HumanWG-6 v3.0 Expression BeadChip kit from the Illumina one-channel system (Illumina, San Diego, CA, USA) was selected.
MATERIALS AND METHODS
Experimental design
In order to carry out a systematic assessment of the impact of co-culture on MSCs, MSCs from six different individuals were used (n = 6), whereas the EC used were pooled from multiple donors. The MSCs were treated with pooled ECs at a ratio of 5 to 1. MSCs from the same donor cultured alone served as control. Two time points, at 5 and 15 days, were chosen based on preliminary kinetic experiments. Each time point comprised six samples in the co-culture group (T) and six in the control group (C) yielding 24 samples in all. The MSC samples were sourced from different donors and denoted as D1 to D6. The sample from donor X in the co-culture group was paired with the sample from donor X in the control group.
The microarray platform chosen was Illumina, a one-channel system. The HumanWG-6 v3.0 Expression BeadChip system (Illumina, San Diego, CA, USA) was used. The single channel platform provides the simple design of 1 sample hybridized to 1 array on a microarray slide. This particular chip has six arrays on each slide.
Cell culture and characterization
The first passage of primary human MSCs were purchased from StemCell Technologies and cultured in MesenCult® complete medium (StemCell Technologies, Vancouver, BC, Canada). MSCs were characterized by flow cytometry, which showed that more than 90% of the cells expressed surface markers of CD44, CD29, CD105, CD166; less than 1% of the cells expressed CD34, CD14, and CD45. Human umbilical vein endothelial cells (HUVECs) were purchased from Lonza (Clonetics™, Walkersville, MD, USA). Following the manufacturer’s instructions, ECs were cultured and expanded in EGM® Medium (Clonetics™). For both cell types, under passage 4 were used. ECs and MSCs were co-cultured and plated in a 6-well plate (Nunclon, Roskilde, Denmark) at a ratio of 1:5. The cell loading density was 2 × 103/cm2 ECs and 1 × 104/cm2 MSCs in a mixed medium by 5:1 MesenCult® complete medium/EGM medium. As a control group, MSCs were seeded onto 6-well plates in the same medium. The culture medium of both groups was changed twice a week.
Due to the different adherent properties of MSCs and ECs, ECs can be separated by two-step trypsinization. After cultivation for 5 and 15 days, ECs in the co-culture group were removed by treatment with 1 mL trypsin/EDTA which is containing 0.5 mg/mL trypsin and 0.2 mg/mL EDTA (Clonetics®, Walkersville, MD), for 5 min at 37°C. Detached ECs were removed with a phosphate buffered saline (PBS) wash; MSC controls were treated similarly. Finally, the MSCs from both groups were rinsed with 1 mL trypsin for 5 min at 37°C. Following a 5 min centrifugation at 1000 g, 37°C, MSC were collected from test and control groups. The cell pellets were stored at −80°C until RNA extraction. The culture medium was collected for calcium assay and vascular endothelial growth factor vascular endothelial growth factor vascular endothelial growth factor (VEGF) release.
After the separation by trypsinization, the purity of MSCs was tested by flow cytometry (FCM). Phycoerythrin (PE) conjugated CD 31 (Santa Cruz, CA, USA) was used to detect ECs. A flow cytometer and software (BD Accuri™ C6 and BD Accuri CFlow® Plus, BD Biosciences, San Jose, CA, USA) were used for analysis. The positive events (cells) were analyzed as the percentage of gated populations and a histogram was compared with the relevant control.
Immunostaining
After 5 and 15 days of culture, cells grown on coverslips of 18 mm diameter (VWR international, West Chester, PA, USA) were rinsed in PBS for three times and then fixed with 4% paraformaldehyde for ten min at room temperature. To distinguish the two cell types, MSC was labeled with FITC conjugated CD90 (BD Pharmingen™, San Jose, CA, USA) and EC was labeled with Tritc-UEA-1 (Vector Laboratories, Burlingame, CA, USA). The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes™, Waltham, MA, USA) solution and washed three times with PBS. The samples were then mounted and imaged by Nikon 80i fluorescence microscopy (Nikon, Tokyo, Japan).
Total RNA preparation and quality control
Total RNA was isolated from 5-day-old and 15-day-old cultures using the E.Z.N.A.™ Tissue RNA isolation kit (Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s protocol. In brief, after cells were disrupted by TRK lysis buffer and homogenized, 70% ethanol was then added to the cleared lysate and applied to a HiBind® RNA spin column placed in a 2 mL collection tube. After centrifugation at 10,000 g for 15 min and washing, on-membrane DNase I digestion was performed and RNA was eluted with nuclease-free water. RNA was quantified using a NanoDrop ND-1000 Spectrophotometer (ThermoScientific NanoDrop Technologies, Wilmington, DE, USA). RNA integrity is a critical step in obtaining high-quality gene expression data. The quality of RNA was analyzed with an Agilent 2100 Bio-analyzer (Agilent Technologies, Santa Clara, CA, USA) and only samples with an RNA Integrity Number (RIN) of ≥7.5 were included in the microarray experiments.
RNA labeling, amplification and microarray hybridization
From each sample, 250 ng of total RNA was reverse-transcribed, amplified and labeled with Biotin-16-UTP using the Illumina TotalPrep RNA Amplification Kit purchased from Applied Biosystems/Ambion (Austin, TX, USA). The amount and quality of the Biotin-labeled cRNA was controlled by both the NanoDrop® spectrophotometer and the Agilent 2100 Bioanalyzer. The Illumina TotalPrep RNA Amplification Kit generated biotinylated and amplified RNA for hybridization with Illumina Sentrix arrays. First, an oligo(dT) primer bearing a T7 promoter in combination with a reverse transcriptase synthesize a first strand full-length cDNA. The cDNA then undergoes second-strand synthesis, and becomes a template for further transcription by T7 RNA polymerase. Thousands of biotinylated cRNAs are generated in each sample after the in vitro transcription.
The biotin-labelled cRNA was then hybridized to the HumanWG-6 v3.0 Expression BeadChip which can target more than 48,000 probes from the human genome database. Hybridization was performed following the Whole-Genome Gene Expression Direct Hybridization Assay Guide from Illumina: 1500 ng cRNA was hybridized at 58°C for 17 hours. By adding streptavidin-Cy3 following the hybridization and washing, the signals of arrays were detected by the Illumina iScan Reader.
Data normalization and quality control
For quality control, the data from the scanning of arrays on the Illumina iScan Reader were evaluated by GenomeStudio and J-Express 2009 (8) (Bergen, Norway; http://jexpress.bioinfo.no/site/, accessed May 22, 2013). After scanning, the raw data were imported into GenomeStudio and several different quality control steps were undertaken. There were 7 control categories built into the hybridization assay system. These covered every aspect of an array experiment, from the biological specimen to sample labeling, hybridization, and signal generation. The GenomeStudio application automatically tracks the performance of these controls and generates a report for each array in the matrix. The full range of technical controls in GenomeStudio confirmed that all the samples were of high quality. All samples had a signal-to-noise ratio well above the threshold value of 10. Before they were compiled into an expression profile data matrix, the arrays within each experiment were quantile-normalized to allow them to be compared.
Microarray data analysis
The SampleProbeProfile was loaded into J-Express 2009 as two separate experiments: 5-day and 15-day. A box plot is one means of visualizing the distribution of intensities for all arrays. These distributions need to be similar for the different samples to be comparable. Samples which behave differently from the other samples, regardless of biology are denoted as outliers and can be detected by clustering and/or projection: these two methods can also be applied to detect batch effects.
To identify differentially expressed genes between two groups, J-express 2009 software was used for significance analysis of microarrays (SAM) (9). SAM is a statistical technique for finding significant gene changes in a set of microarray experiments. Since a donor-paired group was used in this experiment, gene expression measurements were analyzed by a paired SAM method. SAM computed a statistic di for each gene i to indicate the strength of the relationship between gene expressions and treatment. Repeated permutations of the data were performed to determine whether expression of any genes was significantly related to the co-culture treatment. In the current study, changes in gene-expression profile were identified with an estimated false discovery rate (FDR) of less than 1%. Genes with a fold change of greater than 2 were selected. Differentially expressed genes were mapped to a gene ontology (GO)-directed acyclic graph in J-express 2009 and compared with the total number of genes to determine the over-representation of GO terms.
Identification of significantly overrepresented functions
In order to interpret and classify each cluster in more detail by ontological properties, lists of overrepresented genes from the SAM analysis were submitted to the Database for Annotation, Visualization and Integrated Discovery (DAVID) (10, 11). The clusters of genes with significantly similar ontology were generated by the Gene Functional Classification tool in DAVID when comparing inputted gene lists against the whole gene list of the Human Genome. In the present study, the stringency of medium was selected to yield a set of ontological groups. The genes of similar functions were grouped through the DAVID bioinformatics database (http://david.abcc.ncifcrf.gov, accessed May 22, 2013), a classification system intended to identify over-represented biological processes and key pathways.
Validation microarray data by RT-PCR
In order to validate the microarray data, quantitative real time PCR was performed on selected up- and down-regulated genes, using TaqMan pre-developed gene expression assays: Hs00863478_g1 (FLG), Hs00362607_m1 (CD 93), Hs00173787_m1 (CALCRL), Hs00169795_m1 (VWF), Hs01029142_m1 (ALP), Hs00231692_m1 (RunX2), Hs00164099_m1 (Col IA2), and TaqMan GAPDH (4333764T). cDNA that was reverse transcripted from 1000 ng total RNA was used in 96-well plate PCR reaction. Mixtures were made up in 10 μL volume and triplicates of each sample for each target gene. Amplification was carried out in 96-well plates on a StepOnePlus PCR system (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s recommendations. Gene expression was determined by the comparative Ct method, normalizing expression to the reference gene GADPH.
Calcium content and VEGF release
Calcium release was measure by using a Fluo-4 Direct™ Calcium Assay kit (Molecular Probes, Carlsbad, CA, USA; Cat. no. F10471). In brief, the working solution was prepared according the instructions and added to cells directly. After cells incubated at room temperature for 30 min, the fluorescence intensity was measured with plate reader FLUOStar OPTIMA at 485 nm excitation and emission at 520 nm. VEGF concentrations in the supernatant of culture medium were determined by using a human VEGF ELISA Kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. Triplicate measurements were performed for two assays.
Statistical analysis
For the microarray study, the SAM method was used for identification of differentially expressed genes. Other statistical analysis was performed by Student’s t test. Results are expressed as mean ± SEM. A value of p<0.05 was considered significant.
RESULTS
Cell culture and characterization
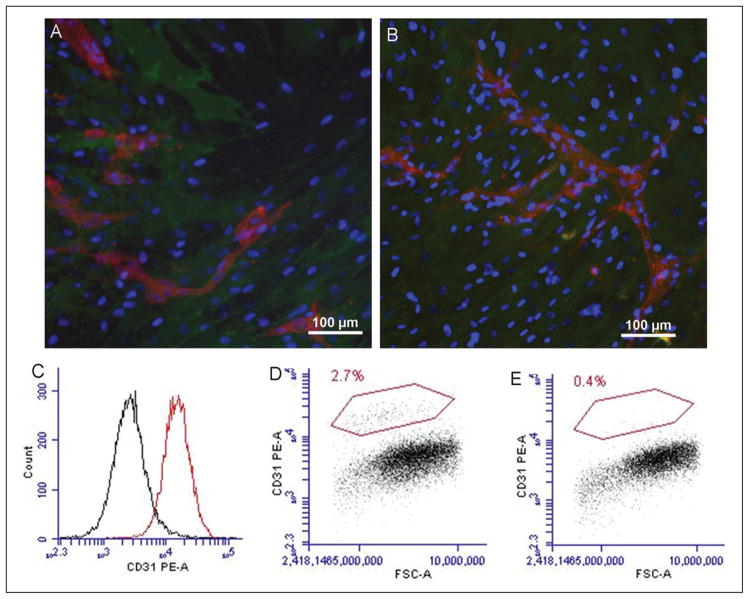
To investigate morphology of ECs and MSCs after co- culturing, fluorescent images were obtained. Images revealed that ECs were able to form chains and capillary-like networks among MSCs after 5 days (Fig. 1A) and 15 days (Fig. 1B). To assess the effect of trypsinization, PE-conjugated CD31 was first tested with ECs (Fig. 1C); a high expression of CD31 on ECs showed reliable efficiency of the antibody. Co-cultured MSCs were then incubated with the PE-CD31 after removing ECs. The results indicated that the amount of remaining ECs was less than 3% after 5 days (Fig. 1D) and less than 1% after 15 days (Fig. 1E).
Fig. 1.
Cell morphology after co-culturing and detection of ECs by FCM. A, B: Morphology of co-culturing, endothelial cells (ECs) was labeled with Tritc-UEA-1 (red) and mesenchymal stem cells (MSCs) with FITC-CD90 (green), the nuclei (blue) were stained with DAPI. ECs were able to form chains and capillary-like networks among MSCs after 5 days (A) and 15 days (B). C: CD31 was highly expressed in ECs. The remaining amount of ECs in co-cultured MSCs was less than 3% at 5 days (D) and less than 1% at 15 days (E) after trypsinization treatment.
Global visualization analysis by Hierarchical Clustering and Correspondence Analysis plots
Before differential expression analysis, a global visualization of the whole experiment can be useful for the subsequent analysis and quality control. In all, 48,803 genes were identified from the whole data set. After quantile-normalization, all arrays within each experiment had the same distribution of data and could now be compared.
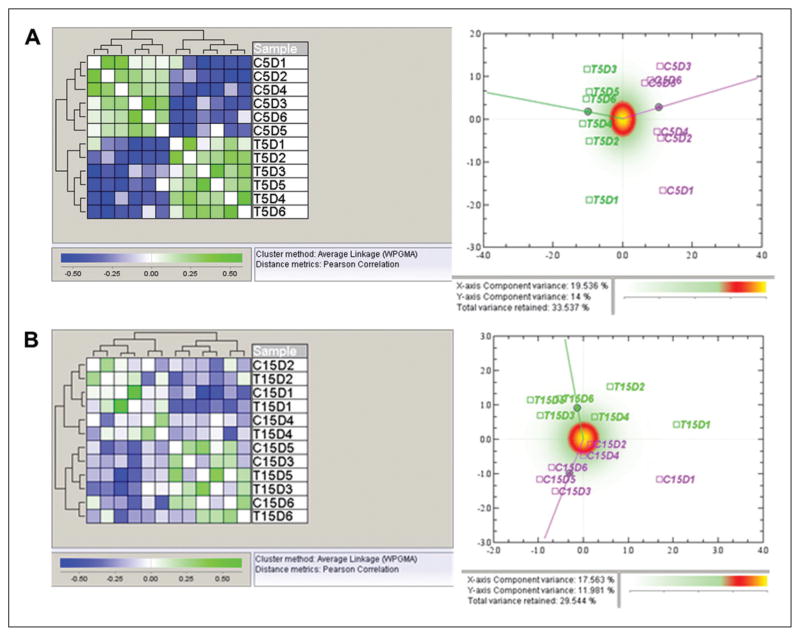
The global effects at the two different time points were investigated in two different ways: Correspondence Analysis (CA)-plots (Fig. 2, left) and Hierarchical Clustering (HC) (Fig. 2, right). In the 5-day experiment, two main clusters were revealed. The two clusters correspond to the co- culture (T) and control (C) groups, meaning that the similarity between the two groups is greater than that among the different donors. This was also confirmed by the clear difference in the CA plot. However, in the 15-day experiment, the two main clusters do not always correspond to the co-cultured treatment (except for donors 3 and 5), but rather show greater similarity to the samples from donors 1, 2, and 4 than between the co-culture and control groups. In the CA plot for the 15-day experiment, the data show a difference between the two groups, but not as distinct as in the 5-day experiment. This result indicates that ECs had larger genotype effects on MSCs after 5 days of co-culture than after 15 days. Both the 5-day and 15-day experiments showed that the data set was of good quality and no outliers were observed.
Fig. 2.
Global views of whole data set. A: Hierarchical clustering (left) and correspondence analysis plot (right) of samples from the quantile-normalized data from the 5-day experiment. In the hierarchical clustering of normalized data using Pearson correlation as distance measure, there were two main clusters which correspond to the co-culture (T) and control (C) groups. In the CA plots, the control group is displayed in pink, and the co-culture group in green. In the correspondence analysis plot for the 5-day experiment, there is a distinct separation between the control group and the co-culture group, which indicates a differential expression of genes on the global level. B: Hierarchical clustering (left) and correspondence analysis plot (right) of samples from the quantile-normalized data from the 15-day experiment. Hierarchical clustering using Pearson correlation as distance measure gives two main clusters. These clusters do however not correspond to the biological groups, but rather show that the similarity between the samples from the same donor is larger than the similarity between the co-culture and control groups. In the correspondence analysis plot for the 15-day experiment, data show a separation between the control group and the co-culture group, but not as distinct as in the 5-day experiment.
Differential expression analysis
In order to list the genes differentially expressed between the two groups, the 48,803 genes were analyzed by SAM. Comparisons of gene expression between the control and co-cultured groups showed 285 genes and 77 genes with FDR less than 1% and fold-changes greater than 2 in the 5-day and 15-day experiments, respectively. Table I shows the top differentially regulated genes; that is, these are the genes that underwent the most pronounced changes from the whole genotype. The top gene lists for the two different time points showed similar trends and shared some of the overrepresented genes (Tab. I).
TABLE I.
TOP 20 DIFFERENTIALLY REGULATED GENE LIST
| 5 days | 15 days | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| PROBE_ID | Gene symbol | Fold Change | FDR (i) | PROBE_ID | Gene Symbol | Fold Change | FDR (i) |
| Up-regulated | Up-regulated | ||||||
|
| |||||||
| ILMN_1752755 | VWF | 100.664 | 0 | ILMN_1752755 | VWF | 62.564 | 0 |
| ILMN_1704730 | CD93 | 23.926 | 0 | ILMN_1704730 | CD93 | 15.476 | 0 |
| ILMN_1660114 | MMRN1 | 14.994 | 0.237 | ILMN_1812968 | SOX18 | 8.798 | 0 |
| ILMN_2071809 | MGP | 13.798 | 0.201 | ILMN_1719236 | CDH5 | 8.377 | 0 |
| ILMN_1719236 | CDH5 | 12.865 | 0 | ILMN_2142185 | CLEC14A | 5.505 | 0 |
| ILMN_1812968 | SOX18 | 11.923 | 0 | ILMN_1806733 | COL18A1 | 4.625 | 0 |
| ILMN_1778444 | FKBP5 | 11.398 | 0 | ILMN_1660114 | MMRN1 | 4.55 | 0 |
| ILMN_2142185 | CLEC14A | 10.417 | 0 | ILMN_1732799 | CD34 | 4.543 | 0 |
| ILMN_1651958 | MGP | 10.047 | 0.21 | ILMN_1689518 | PECAM1 | 4.398 | 0 |
| ILMN_2184373 | IL8 | 9.954 | 0 | ILMN_2413158 | PODXL | 4.199 | 0 |
| ILMN_2313672 | IL1RL1 | 9.2 | 0.224 | ILMN_1653466 | HES4 | 4.18 | 0 |
| ILMN_1748473 | GIMAP4 | 7.819 | 0.282 | ILMN_2341229 | CD34 | 4.023 | 0 |
| ILMN_1701603 | ALPL | 7.811 | 0.655 | ILMN_1796288 | COL5A3 | 3.706 | 0 |
| ILMN_1675453 | HHIP | 7.166 | 0.333 | ILMN_1668092 | ESAM | 3.671 | 0 |
| ILMN_1658494 | C13orf15 | 6.481 | 0 | ILMN_1752932 | MPZL2 | 3.511 | 0 |
| ILMN_1666733 | IL8 | 6.44 | 0 | ILMN_1728785 | GPR116 | 3.498 | 0 |
| ILMN_1663640 | MAOA | 6.386 | 0 | ILMN_1748473 | GIMAP4 | 3.371 | 0 |
| ILMN_1754538 | C10orf58 | 6.334 | 0.661 | ILMN_2212878 | ESM1 | 3.368 | 0 |
| ILMN_1689518 | PECAM1 | 6.288 | 0 | ILMN_1654324 | HEYL | 3.023 | 0 |
| ILMN_2411236 | NRCAM | 5.641 | 0 | ILMN_1757440 | FAM69B | 2.936 | 0 |
| ILMN_1707727 | ANGPTL4 | 5.569 | 0 | ILMN_2371055 | EFNA1 | 2.884 | 0 |
|
| |||||||
| Down-regulated | Down-regulated | ||||||
|
| |||||||
| ILMN_2134130 | FLG | −21.279 | 0 | ILMN_2134130 | FLG | −3.333 | 0 |
| ILMN_1813704 | KIAA1199 | −12.87 | 0 | ILMN_1813704 | KIAA1199 | −2.725 | 0 |
| ILMN_1803213 | MXRA5 | −9.256 | 0 | ILMN_1678812 | HAPLN1 | −2.713 | 0 |
| ILMN_1700081 | FST | −8.503 | 0 | ILMN_1791447 | CXCL12 | −2.65 | 0 |
| ILMN_1810172 | SFRP4 | −8.081 | 0.187 | ILMN_2304512 | SAA1 | −2.579 | 0 |
| ILMN_1730777 | KRT19 | −7.823 | 0 | ILMN_1689111 | CXCL12 | −2.453 | 0.837 |
| ILMN_1678842 | THBS2 | −6.88 | 0 | ILMN_1733415 | MFAP5 | −2.424 | 0 |
| ILMN_1720695 | LOC730833 | −6.79 | 0 | ILMN_2376953 | KCNK2 | −2.253 | 0 |
| ILMN_1734653 | FNDC1 | −6.261 | 0 | ILMN_1810628 | KIAA0367 | −2.223 | 0 |
| ILMN_1667692 | PTGIS | −6.095 | 0 | ILMN_1735910 | VMO1 | −2.179 | 0.35 |
| ILMN_2163873 | FNDC1 | −6.079 | 0 | ILMN_1719759 | TNC | −2.163 | 0.814 |
| ILMN_1726711 | PENK | −5.907 | 0 | ILMN_1765990 | KCNK2 | −2.145 | 0 |
| ILMN_2200836 | HSPB7 | −5.584 | 0.267 | ILMN_1656920 | CRIP1 | −2.141 | 0 |
| ILMN_2385672 | ELN | −5.516 | 0 | ILMN_1801205 | GPNMB | −2.056 | 0 |
| ILMN_1677636 | COMP | −5.473 | 0 | ILMN_1805561 | SLC14A1 | −2.02 | 0.715 |
| ILMN_1678812 | HAPLN1 | −5.464 | 0 | ||||
| ILMN_1791447 | CXCL12 | −5.075 | 0 | ||||
| ILMN_1676663 | TNFRSF11B | −5.002 | 0 | ||||
| ILMN_1699651 | IL6 | −4.942 | 0 | ||||
| ILMN_1665035 | KRT14 | −4.862 | 0 | ||||
| ILMN_1736760 | KRT16 | −4.837 | 0.195 | ||||
Classification of genes according to function
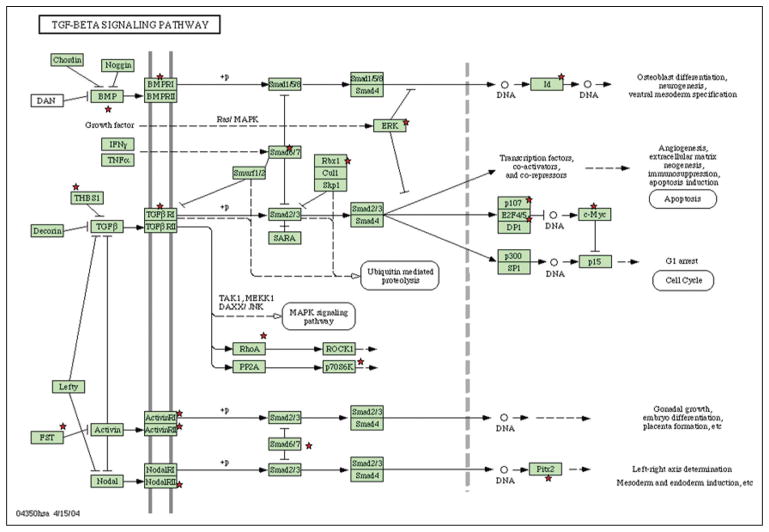
To explore and view functionally related genes together, the two overrepresented gene lists from the 5-day and 15-day experiments were submitted to the DAVID database as separate sets of data. The 5-day gene lists were assessed by Gene Ontology and the pathway database. The data are presented in Table II. In the 5-day experiments, DAVID recognized a total of 218 specific gene symbols out of the list of 285 genes submitted. For the 218 genes identified by the Panther-BP-ALL databank, Gene Ontology (GO) analysis revealed 65 related GO terms. Table II presents the annotations of all the GO terms. The order in the list is based on the gene number involved. Some of the GO terms (Signal peptide, Cell adhesion, Extracellular matrix, Blood vessel development, Cell migration, Response to wounding, ECM-receptor interaction, Mesenchymal cell development, Tube development, etc.) revealed important information about interactions between MSCs and ECs. These indicated that the interactions between the two cell types might go through cell surface receptor-linked signal transduction such as interleukin 6 (IL6), interleukin 8 (IL8), and chemokine (C-X-C motif) ligand 12 (CXCL12). The integrin family – integrin beta 2 (ITGB2), integrin, beta 1 (ITGB1), integrin alpha E (ITGAE), integrin alpha 8 (ITGA8) and integrin alpha V (ITGAV) – are significantly overrepresented in MSCs after being co-cultured with ECs. At the same time, the pathway chart (Tab. 3A) indicated the pathways most involved with the listed genes, such as ECM-receptor interaction, cell adhesion molecules (CAMs), TGF-beta signaling pathway, leukocyte transendothelial migration, and focal adhesion. With respect to bone development, the most interesting finding was that 19 over-represented genes were involved in the TGF-beta pathway (Fig. 3).
TABLE II.
GO TERM ANNOTATION OF 5 DAYS’ GENE LIST
| GO Term | Count | Fold Enrichment | Enrichment Score | P Value |
|---|---|---|---|---|
| Signal peptide | 98 | 2.628575622 | 17.6667 | 1.16E-21 |
| Cell adhesion | 28 | 5.657907013 | 10.1803 | 7.02E-13 |
| Extracellular matrix | 27 | 5.450620082 | 9.15492 | 3.16E-12 |
| Blood vessel development | 18 | 5.183016488 | 6.70772 | 6.57E-08 |
| Cell migration | 17 | 4.362624794 | 5.19 | 1.72E-06 |
| Response to wounding | 22 | 3.059008886 | 3.64 | 9.51E-06 |
| Immunoglobulin domain | 19 | 3.469646478 | 3..29 | 9.94E-06 |
| Hydroxylation | 9 | 11.21025606 | 3.22 | 1.28E-06 |
| EGF-like region, conserved site | 17 | 4.471613194 | 2.94 | 1.35E-06 |
| Chelation | 4 | 41.72706422 | 2.83 | 8.97E-05 |
| Mesenchymal cell development | 6 | 8.439014025 | 2.8 | 6.81E-04 |
| ECM-receptor interaction | 10 | 6.643437863 | 2.58 | 1.36E-05 |
| Domain: CTCK | 4 | 14.25152905 | 2.45 | 0.00262892 |
| Response to organic substance | 25 | 2.480940351 | 2.37 | 6.09E-05 |
| Regulation of locomotion | 11 | 4.168493974 | 2.27 | 3.08E-04 |
| Prostaglandin receptor activity | 3 | 24.53757225 | 2.15 | 0.00613438 |
| Urogenital system development | 8 | 5.087869325 | 2.07 | 9.76E-04 |
| C-type lectin-like | 7 | 5.6740638 | 2.02 | 0.00145361 |
| Tube development | 10 | 3.29450998 | 1.99 | 0.00335422 |
| IL 17 Signaling Pathway | 4 | 10.43809524 | 1.9 | 0.00518607 |
TABLE III.
| A: 5 days
| |||
|---|---|---|---|
| Term | Count | P value | Genes |
| ECM-receptor interaction (hsa04512) | 10 | 1.27E-05 | VWF, SDC1, CD44, ITGA5, LAMA5, ITGA8, COMP, ITGA11, ITGA10, THBS2 |
| Cell adhesion molecules (hsa04514) | 10 | 4.19E-04 | NRCAM, SDC1, CADM1, CD34, ITGA8, PECAM1, CLDN5, ESAM, ITGB2, CDH5 |
| TGF-beta signaling pathway (hsa04350) | 7 | 0.0039436 | ID2, CDKN2B, ID1, COMP, FST, THBS2, BMP6 |
| Leukocyte transendothelial migration (hsa04670) | 7 | 0.0167636 | PECAM1, CLDN5, ESAM, ITGB2, CXCL12, MMP2, CDH5 |
| Focal adhesion (hsa04510) | 9 | 0.0236411 | VWF, ITGA5, LAMA5, ITGA8, COMP, ITGA11, ITGA10, THBS2, MYLK |
| B: 15 days
| |||
|---|---|---|---|
| Term | Count | P value | Genes |
| Cell adhesion molecules (hsa04514) | 6 | 3.80E-04 | NRCAM, CD34, PECAM1, CLDN5, ESAM, CDH5 |
| Notch signaling pathway (hsa04330) | 4 | 0.0015182 | MFNG, NOTCH4, JAG2, JAG1 |
| Leukocyte transendothelial migration (hsa04670) | 5 | 0.0024649 | PECAM1, CLDN5, ESAM, CXCL12, CDH5 |
| ECM-receptor interaction (hsa04512) | 4 | 0.0079119 | VWF, LAMA5, TNC, COL5A3 |
Fig. 3.
Genes involved in the TGF beta signaling pathway after 5 days. After the overrepresented 5-day gene lists were submitted to the DAVID database (http://www.DAVID.org), 19 overrepresented genes were indicated as involved in the TGF-beta pathway as separate sets of data. Those 19 genes were labeled with red stars in this picture.
In the 15-day gene list, 65 specific gene symbols were identified by DAVID from a list of 78 differentiated genes. According to the KEGG-pathway database, the pathway map of the 15-day gene list disclosed four different pathways: cell adhesion molecules (CAMs), notch signaling pathway, leukocyte transendothelial migration, and ECM-receptor interaction (Tab. 3B).
Validation of microarray data by RT-PCR
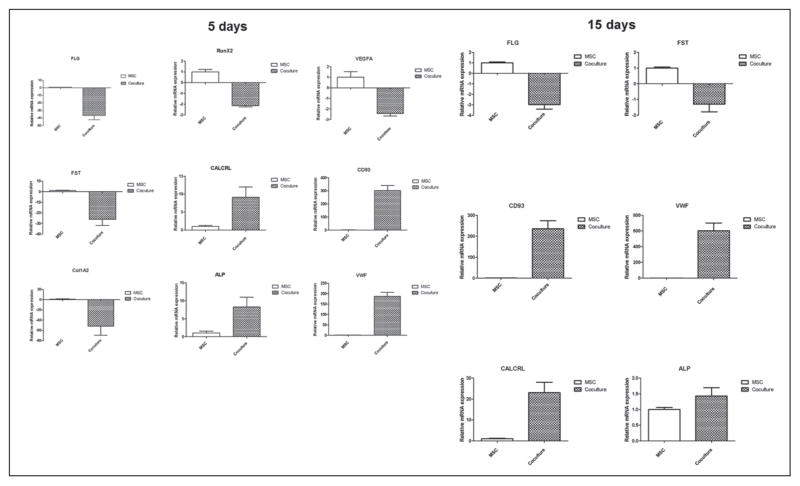
Several selected genes from the overrepresented list under mono-and co-culture conditions for 5 days and 15 days were confirmed by real time PCR. The relative expression levels are represented as MSC co-culture over monoculture (Fig. 4). Thus, the gene expressions identified by microarray analysis were confirmed by a second method: real time PCR.
Fig. 4.
Validations of microarray data by real time RT-PCR. Left: mRNA of MSC with selected up-regulated genes (CALCRL, ALP, vWF, CD 93) and down-regulated genes (FLG, VEGFA, Runx2, FST, Col1a2) from 5-day gene list. Right: mRNA of MSC after 15 days’ co-culturing with selected up-regulated genes (CALCRL, ALP, vWF, CD 93) and down-regulated genes (FLG, VEGFA, FST). Comparing these results with microarray data, they did not have the same values of fold changes but the same tendencies of data were confirmed and validated.
Calcium content and VEGF release
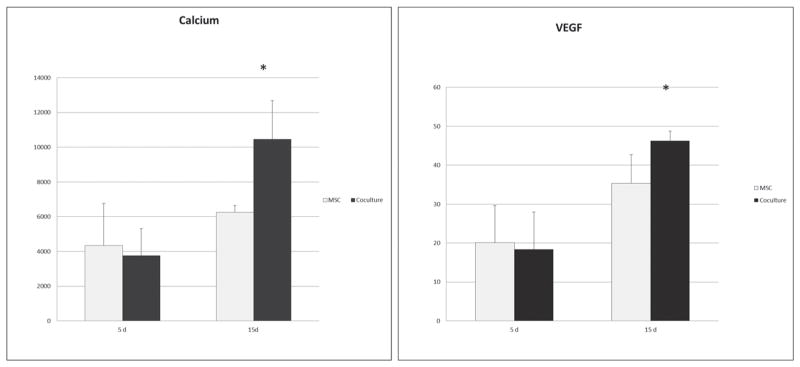
To access the osteogenic differentiation of MSCs after being co-cultured for 5 days and 15 days, intracellular calcium concentration was measured (Fig. 5). The calcium level was detected in both groups and was significantly enhanced by co-culturing after 15 days. Surprisingly, as a critical factor related to angiogenesis and wound healing, VEGF showed a significant increase in co-cultured MSCs after 15 days.
Fig. 5.
Intracellular calcium content and VEGF release. In order to address the bidirectional gene regulation of angiogenesis and osteogenesis by co-culture, an intracellular calcium and VEGF concentration were detected after 5 days and 15 days. Both calcium and VEGF with a significantly higher level after 15 days were determined.
DISCUSSION
Co-culture of MSCs and ECs is a promising strategy for bone tissue engineering (2). However, the biological processes underlying cell-to-cell communication between MSCs and ECs are not well understood. The establishment of heterotypic cell contacts demonstrates that the co-culture system is a useful tool for studying paracrine and/or cell contact-mediated interactions between MSCs and ECs for application in bone tissue engineering. In the present study, the systematic analysis of patterns of global gene expression arising from cross-talk between MSCs and ECs and a map of genes differentially expressed by MSCs after co-culture with ECs for 5 days and 15 days were generated. The dataset further classified the subsets of genes according to function, demonstrating that this classification reflected crucial hallmarks of physiological features of the impact of EC after different periods of co-culture.
Bidirectional gene regulation of angiogenesis and osteogenesis by co-culture
It has been shown that MSC differentiation can be induced to OBs by the addition of dexamethasone, ascorbic acid, and beta-glycerophosphate (12), and can also be induced to form ECs when the culture medium is supplemented with VEGF (13). These studies indicate that an important feature of MSCs is their multi-lineage differentiation potential. Under appropriate inductive conditions, MSCs are able to acquire characteristics of cells such as OBs and ECs. However, the molecular mechanisms that govern MSC differentiation are not fully understood. The present co-culture model simulates in vivo conditions involving natural bone tissue and proved to be a useful model for exploring cellular induction.
Co-culture of ECs and MSCs induced complex, bidirectional gene regulation mechanisms. The top 20 gene list displayed the genes from the whole genotype that had undergone the most pronounced changes after co-culture. As shown in Table I, co-culture resulted in up-regulation of genes related to angiogenesis, such as von Willebrand factor (vWF), platelet/endothelial cell adhesion molecule-1 (PECAM1), cadherin 5 (CDH5), angiopoietin-related protein 4 (ANGPTL4), and cell surface antigen CD34. Also up-regulated in the co-cultured-MSC were markers of osteogenesis, such as alkaline phosphatase (ALP), FK506 binding protein 5 (FKBP5) (14) and bone morphogenetic protein (BMP). The list of overrepresented genes demonstrated the bidirectional gene regulation of angiogenesis and osteogenesis after co-culture. Moreover, the enhanced intra-cellular calcium level and increased VEGF release in the culture medium have strengthened this finding from the protein detection level.
This finding could be a good molecular explanation for the rat study done by Yu et al, in which implantation of a polymer scaffold containing co-cultured ECs and bone marrow-derived OBs resulted in improved osteogenesis and enhanced vascularization (7). It suggests a strategy for simultaneously improving angiogenesis and osteogenesis in engineered bone constructs. However, to apply this for future clinical use, many remaining issues need to be solved such as the proper cell sources for endothelial cells as well as safe and practical protocol of co-culturing during the whole procedure. Of course, if further studies at the functional level can be done to reveal the specific signaling or molecular underlying interactions, then co-culturing may be replaced with direct delivery of a certain protein for wound healing or bone regeneration.
Angiogenic regulation and cell-matrix interactions
Crosstalk between the ECs and MSCs also had a significant impact on cell adhesion and cell-ECM communication. The list of top up-regulated genes also included ECM-related genes, such as clusters of differentiation 93 (CD 93), CDH5, vWF, and multimerin 1 (MMRN1). This trend was also noted in the cluster analysis. In the 5-day experiment, GO clustering (Tab. II) showed obvious changes in signal peptide (98 genes), cell adhesion (28 genes), ECM (27 genes), blood vessel development (18 genes), cell migration (17 genes), and ECM-receptor interaction (10 genes). After 15 days, the enhanced VEGF release in co-culture indicated a clear angiogenesis induction in the microenvironment. These data suggest that cell adhesion and cell-ECM interaction play a role in crosstalk between the two cell types. Extracellular matrix secreted by the cells could provide a local environment for cell-cell interactions and signals driving differentiation. A previous study has shown that MSCs cultured on EC-matrix induced EC differentiation and also indicated that EC-matrix contained certain signals and factors that modified MSC differentiation into ECs (15). The microarray data and the overrepresented expression of angiogenic markers and ECM-related genes after 5 days and 15 days are suggestive of EC-matrix interactions and this conclusion is supported by the present study.
TGF-β pathway might be the mechanism of osteogenic induction by EC
KEGG-Pathway mapping implicated several different pathways, including the TGF-β pathway, which is important for differentiation and cell proliferation in osteogenesis. The transforming growth factor-β (TGF-β) family members include TGF-β subtypes, activins, and BMP, all secreted as cytokines and involved in multifunctional signaling. Interestingly, bone matrix is one of the richest reservoirs of TGF-β and OBs possess several different TGF-β receptors (16). Several studies have indicated that TGF-β regulates OB functions through integrin receptors (16–18). TGF-β family members bind to the Type II receptor and simultaneously recruit Type I receptors to form a heteromeric complex (16). In the 5-day co-culture group, the following 7 genes were recruited to this pathway (Fig. 3): BMP, follistatin (FST), v-myc myelocytomatosis viral related oncogene (c-Myc), inhibitor of DNA binding (Id), extracellular signal-regulated kinase (ERK), transcription factor (DP1), and cartilage oligomeric matrix protein (THBS1). FST is a natural antagonist of activin, so the ratio of activin and FST is crucial for osteoblast differentiation (19). The down-regulated FST expression in co-cultured MSCs indicated this stimulation effect by ECs might be activin-dominated, which is similar to BMP 2-induced osteoblast differentiation. These findings are consistent with previous studies, in which co-culture showed increased expression of osteoblast marker ALP and positive proliferation (5, 6), suggesting that ECs direct MSCs toward an OB phenotype, which may allow them to be regarded as osteoinductive mediators (2). However, only early bone marker was up-regulated, which indicated that ECs might transiently retard osteogenic differentiation of MSCs and keep them in a pre-OB stage. Moreover, the mechanism of this controlling could be due to suppression of the TGF-β pathway. Previous study done by Kyoko et al (20) showed that TGF-β suppressed MSCs differentiation into terminal OBs and that this suppression could be reversed by adding the TGF-β inhibitors SB431542 and Ki26894.
In summary, gene profiling generated by microarray disclosed a complex interaction between ECs and MSCs. A wide range of MSC genes and biological processes was significantly influenced by the addition of less than 20% ECs. These results may contribute to better understanding of bone physiology. In particular, the bidirectional gene regulation of angiogenesis and osteogenesis under co-culture conditions suggests new avenues for controlling and modulating bone-healing in tissue engineering, through cell-to-cell interactions. Although several critical issues regarding the standardization of Illumina BeadChips such as RNA quality control, six replica of each group, and normalization of the internal control have been addressed here, there are still some limitations regarding this technique. For example, each microarray can only provide analysis of the genes that are included on the array, and every gene annotated in the NCBI database is certainly not included in the design. Despite the full discovery of the human-genome project, most genes still constitute the dark matter of the body. In order to determine the mechanisms underlying the bidirectional process as well as the gene profiling generated from this study, other functional assays combined with the proteomic method are needed.
CONCLUSIONS
This study has generated a global map of gene expressions of the interaction between MSCs and ECs in a direct-contact model, by which to determine potential, relative, functional processes and pathways. The results showed a significant impact of ECs on MSCs after 5 days and 15 days of co-culture, especially with respect to bidirectional gene regulation of angiogenesis and osteogenesis, through cell signaling, cell adhesion, and cellular matrix. Cell-matrix interactions and TGF-β signal pathways are implicated in EC-induced gene regulation of MSCs. Further analysis of the microarray data is warranted to explore these cellular and molecular interactions.
Acknowledgments
The authors would like to thank Kjell Petersen, Anne-Kristin Bergen, Norway, for their assistance with J-express software and helpful discussions on bioinformatics. The authors also would like to thank Solveig Angelskår and Rita Holdhus at the NMC Core Facility Laboratory, Bergen, Norway, for technical assistance with the microarray procedures, and Dr. Joan Bevenius for linguistic revisions of the manuscript.
Financial Support: This project was supported by the VascuBone project, European Union FP7; grant number: 242175; The Research Council of Norway ((Stem Cell 180383/V40), (FRIMED, 17734/V50), (156744)); and USPHS grant DE 19938 (TVD).
Footnotes
Conflict of Interest Statement: The authors have declared that no competing interests exist.
References
- 1.Kneser U, Schaefer DJ, Polykandriotis E, Horch RE. Tissue engineering of bone: the reconstructive surgeon’s point of view. J Cell Mol Med. 2006;10(1):7–19. doi: 10.1111/j.1582-4934.2006.tb00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grellier M, Bordenave L, Amédée J. Cell-to-cell communication between osteogenic and endothelial lineages: implications for tissue engineering. Trends Biotechnol. 2009;27(10):562–571. doi: 10.1016/j.tibtech.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Kanczler JM, Oreffo RO. Osteogenesis and angiogenesis: the potential for engineering bone. Eur Cell Mater. 2008;15:100–114. doi: 10.22203/ecm.v015a08. [DOI] [PubMed] [Google Scholar]
- 4.Xing Z, Xue Y, Dånmark S, et al. Effect of endothelial cells on bone regeneration using poly(L-lactide-co-1,5-dioxe-pan-2-one) scaffolds. J Biomed Mater Res A. 2011;96(2):349–357. doi: 10.1002/jbm.a.32989. [DOI] [PubMed] [Google Scholar]
- 5.Villars F, Guillotin B, Amédée T, et al. Effect of HUVEC on human osteoprogenitor cell differentiation needs heterotypic gap junction communication. Am J Physiol Cell Physiol. 2002;282(4):C775–C785. doi: 10.1152/ajpcell.00310.2001. [DOI] [PubMed] [Google Scholar]
- 6.Xue Y, Xing Z, Hellem S, Arvidson K, Mustafa K. Endothelial cells influence the osteogenic potential of bone marrow stromal cells. Biomed Eng Online. 2009;8:34. doi: 10.1186/1475-925X-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, VandeVord PJ, Mao L, Matthew HW, Wooley PH, Yang SY. Improved tissue-engineered bone regeneration by endothelial cell mediated vascularization. Biomaterials. 2009;30(4):508–517. doi: 10.1016/j.biomaterials.2008.09.047. [DOI] [PubMed] [Google Scholar]
- 8.Dysvik B, Jonassen I. J-Express: exploring gene expression data using Java. Bioinformatics. 2001;17(4):369–370. doi: 10.1093/bioinformatics/17.4.369. [DOI] [PubMed] [Google Scholar]
- 9.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(9):R60. [PubMed] [Google Scholar]
- 11.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 12.Jaiswal N, Haynesworth SE, Caplan AI, Bruder SP. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J Cell Biochem. 1997;64(2):295–312. [PubMed] [Google Scholar]
- 13.Oswald J, Boxberger S, Jørgensen B, et al. Mesenchymal stem cells can be differentiated into endothelial cells in vitro. Stem Cells. 2004;22(3):377–384. doi: 10.1634/stemcells.22-3-377. [DOI] [PubMed] [Google Scholar]
- 14.Liu TM, Martina M, Hutmacher DW, Hui JH, Lee EH, Lim B. Identification of common pathways mediating differentiation of bone marrow- and adipose tissue-derived human mesenchymal stem cells into three mesenchymal lineages. Stem Cells. 2007;25(3):750–760. doi: 10.1634/stemcells.2006-0394. [DOI] [PubMed] [Google Scholar]
- 15.Lozito TP, Taboas JM, Kuo CK, Tuan RS. Mesenchymal stem cell modification of endothelial matrix regulates their vascular differentiation. J Cell Biochem. 2009;107(4):706–713. doi: 10.1002/jcb.22166. [DOI] [PubMed] [Google Scholar]
- 16.Nesti LJ, Caterson EJ, Li WJ, et al. TGF-beta1 calcium signaling in osteoblasts. J Cell Biochem. 2007;101(2):348–359. doi: 10.1002/jcb.21180. [DOI] [PubMed] [Google Scholar]
- 17.Dallas SL, Sivakumar P, Jones CJ, et al. Fibronectin regulates latent transforming growth factor-beta (TGF beta) by controlling matrix assembly of latent TGF beta-binding protein-1. J Biol Chem. 2005;280(19):18871–18880. doi: 10.1074/jbc.M410762200. [DOI] [PubMed] [Google Scholar]
- 18.Lee JY, Kim KH, Shin SY, et al. Enhanced bone formation by transforming growth factor-beta1-releasing collagen/chitosan microgranules. J Biomed Mater Res A. 2006;76(3):530–539. doi: 10.1002/jbm.a.30434. [DOI] [PubMed] [Google Scholar]
- 19.Beites CL, Hollenbeck PL, Kim J, Lovell-Badge R, Lander AD, Calof AL. Follistatin modulates a BMP autoregulatory loop to control the size and patterning of sensory domains in the developing tongue. Development. 2009;136(13):2187–2197. doi: 10.1242/dev.030544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi K, Abe M, Hiasa M, et al. Tgf-Beta inhibition restores terminal osteoblast differentiation to suppress myeloma growth. PLoS One. 2010;5(3):e9870. doi: 10.1371/journal.pone.0009870. [DOI] [PMC free article] [PubMed] [Google Scholar]