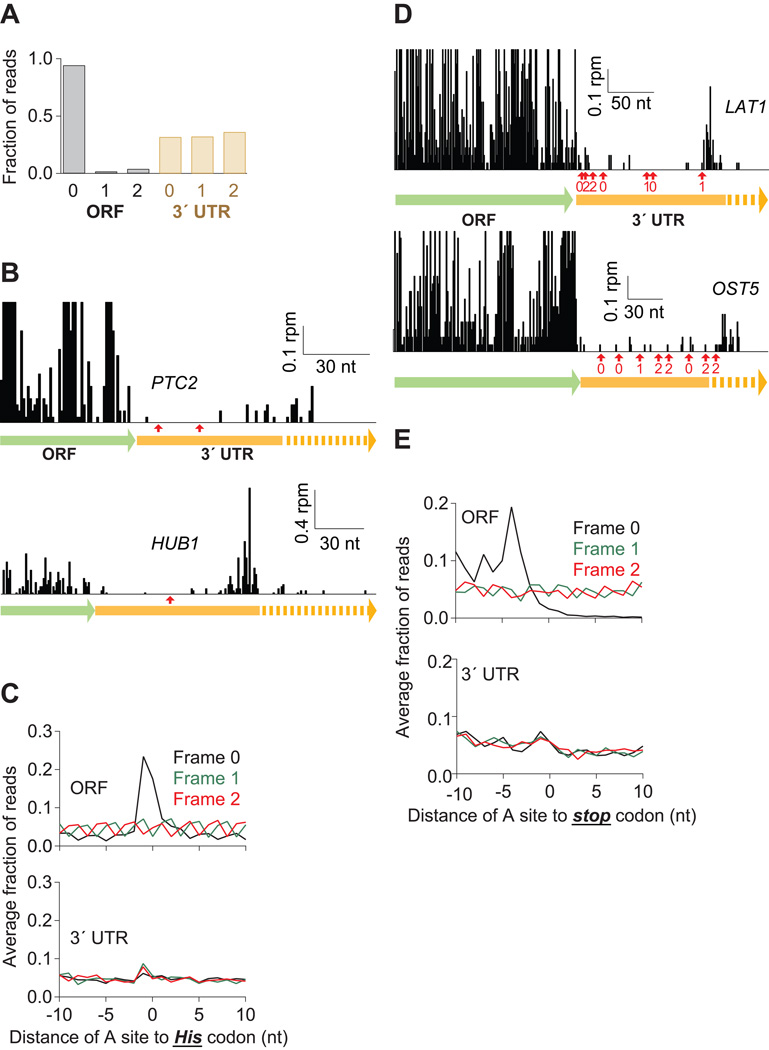
Figure 4. Many Ribosomes Do Not Translate the 3´ UTR.
(A) Fraction of 28-nt reads that map (without mismatches) to each reading frame in the ORF and 3´ UTR for dom34Δ cells. Normalized data were averaged from ribosomes stabilized with CHX, 10 mM MgCl2, or no extra additive, but not GMP-PNP as frame was less well preserved.
(B) Examples of mapped footprints from dom34Δ cells treated with 3-AT on genes (PTC2 and HUB1) with in-frame His codons (indicated with red arrows) in the 3´ UTR. Note that the vertical axis scaling truncates the upper range of ORF reads for PTC2.
(C) Average fraction of ribosome occupancy within a 20-nt window around His codons in a given frame. Data are shown for all three reading frames in ORFs and 3´ UTRs from dom34Δ cells treated with 3-AT.
(D) Examples of mapped footprints from dom34Δ cells on genes (LAT1 and OST5) with stop codons in all three reading frames of the 3´ UTR. Stop codons and their respective reading frame before the poly(A) tail are indicated (red arrows and values 0–2). Note that the vertical axis scaling truncates the upper range of ORF reads.
(E) Average fraction of ribosome occupancy within a 20-nt window around the first occurrence of a stop codon in a given frame. Data are shown for all three reading frames in ORFs and 3´ UTRs in dom34Δ cells.