Figure 5. Ribosome Occupancy in 3´ UTR is Modulated by a Suppressor tRNA or Addition of Diamide.
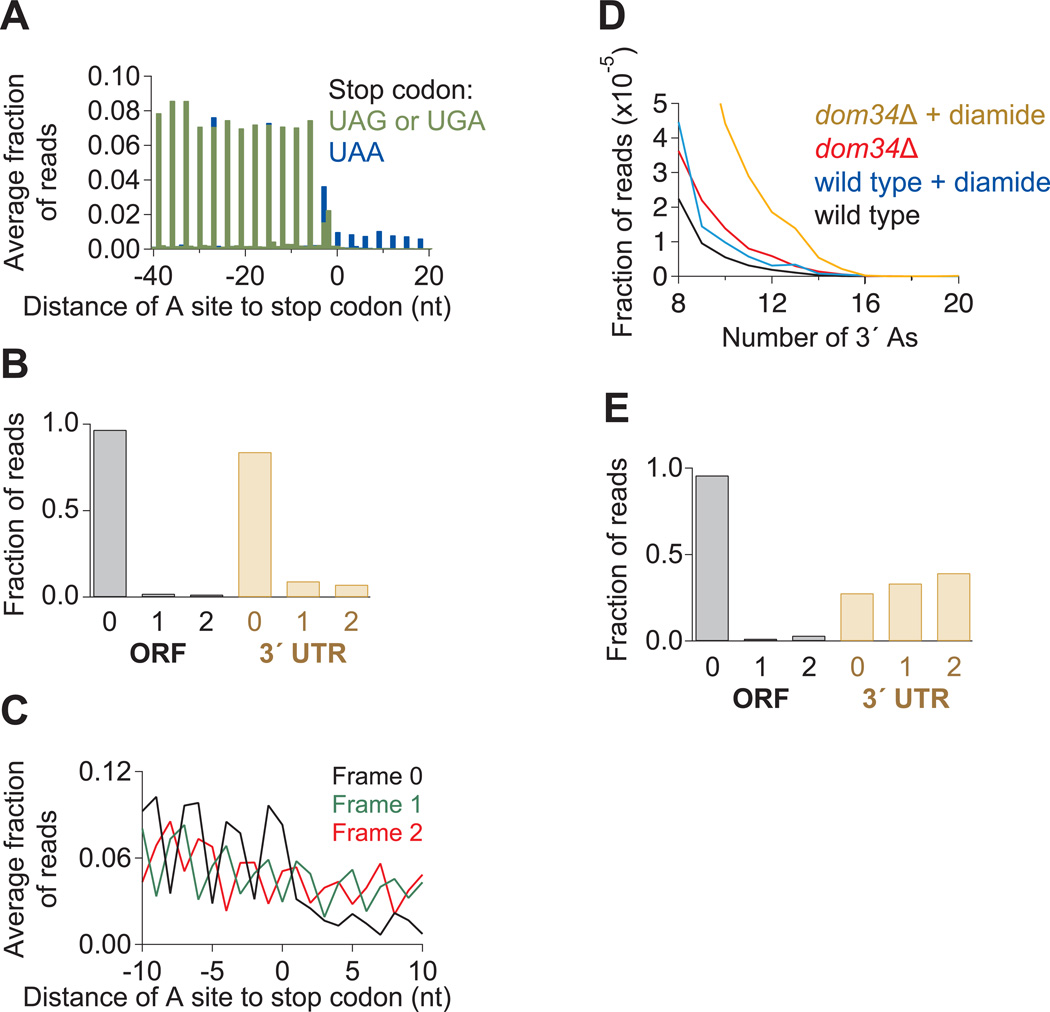
(A) Average fraction of ribosome occupancy in a region around the stop codon of annotated ORFs. Data taken from ski2Δ cells transformed with the SUP4-o tRNA. Only reads that measured 28 nt in length and mapped without mismatches are plotted. Readthrough is increased only when the stop codon is UAA.
(B) Fraction of 28-nt reads that map (without mismatches) to each reading frame in the ORF and 3´ UTR for ski2Δ cells transformed with the SUP4-o tRNA.
(C) Average fraction of ribosome occupancy within a 20-nt window around the first occurrence of a stop codon in a given frame. Data are shown only in 3´ UTRs that follow ORFs terminating with a UAA stop codon. Data from ski2Δ cells transformed with the SUP4-o tRNA.
(D) Histogram of A count on the 3´ ends of footprint reads before alignment to the genome. Data for wild-type and dom34Δ yeast in the presence and absence of diamide are shown.
(E) Fraction of 28-nt reads that map (without mismatches) to each reading frame in the ORF and 3´ UTR for dom34Δ cells treated with diamide.