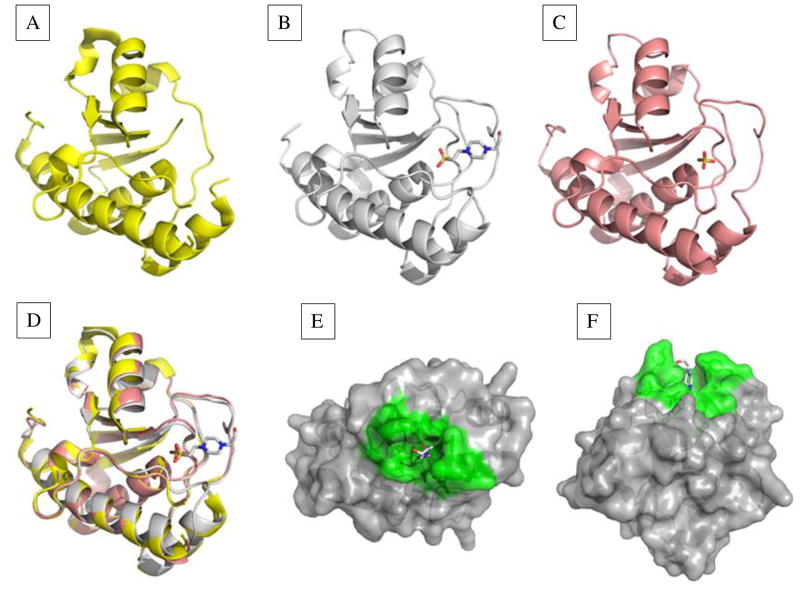
Fig. 1. Structures of the E. histolytica LMW-PTP with no bound ligand, substrate mimic bound, or product mimic bound.
Structures were solved for each given resolution. (A) The apoenzyme structure of 2.2 Å showed a disordered active site (P- loop) (PDB: 3ILY). (B) Two structures, of 2.2 Å and 1.95 Å (PDB: 3IDO; PDB: 3JS5) were obtained with substrate mimic HEPES bound. The P-loop and interacting residues were well-ordered. (C) Structure with the product mimic sulfate (PDB: 3JVI) bound in the active site (1.8 Å). This structure had a well-ordered P-loop, but additional residues on the surface of the active site were disordered. (D) A triple overlay image of structures in (A), (B), and (C) showed changes in enzyme conformation when no substrate, substrate, or product was bound. (E) side view and (F) top view of the LMW-PTP structure with HEPES bound in the active site. Active site and contributing residues are green, and other residues are gray.