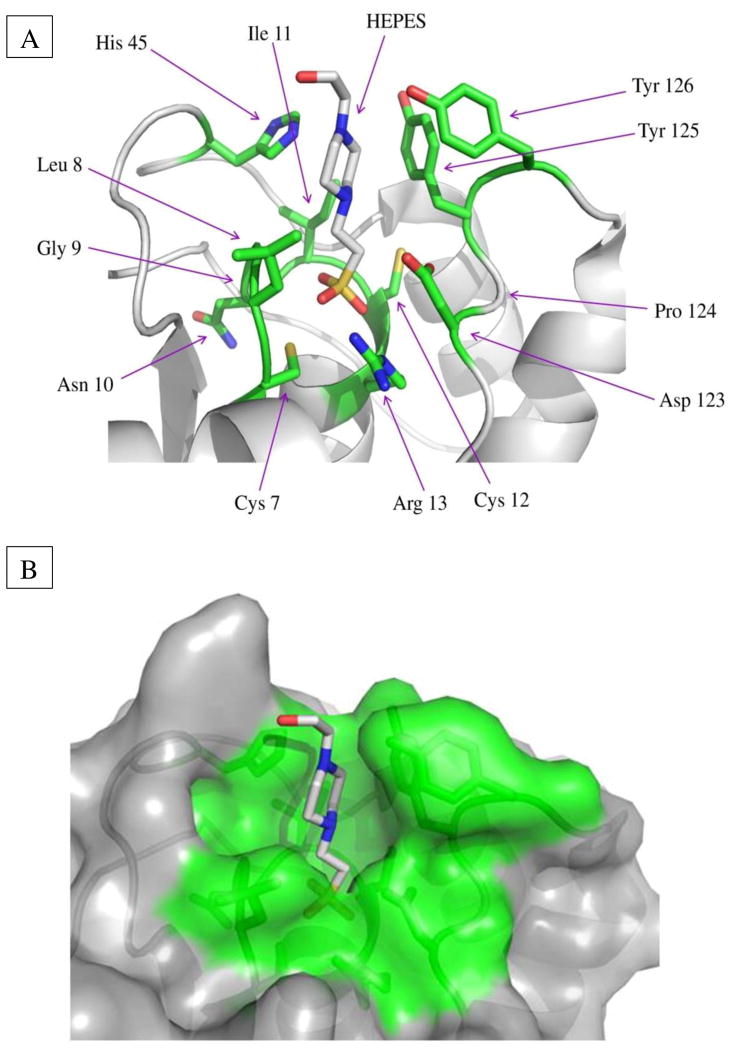
Fig. 2. Active site structure and residues of the E. histolytica LMW-PTP.
(A) In the ribbon model with the substrate HEPES bound, active site residues are green, sulfur atoms yellow, nitrogen atoms blue, and oxygen atoms red. Residues are numbered according to the E. histolytica LMW-PTP protein sequence. Arg, arginine; Asn, asparagine; Asp, aspartic acid; Cys, cysteine; Gly, glycine; His, histidine; Ile, isoleucine; Leu, leucine; Pro, proline; Tyr, tyrosine. Key residues apart from the Cys7 catalytic residue include Cys12, which can form a disulfide bond with Cys7 to prevent irreversible oxidation of Cys7 in the presence of reactive oxygen species, and Arg13, which stabilizes the transition state via hydrogen bonding. The surface loop contains the conserved Asp123, which functions as a general acid/base in catalysis, as well as Tyr125 and Tyr126 residues, which can be phosphorylated by Src family kinases in mammalian LMW-PTPs to regulate enzymatic activity. The His45 residue appears to function in substrate specificity recognition. (B) In the space-filling model, active site and contributing residues are green; residues not contributing to catalysis are gray.