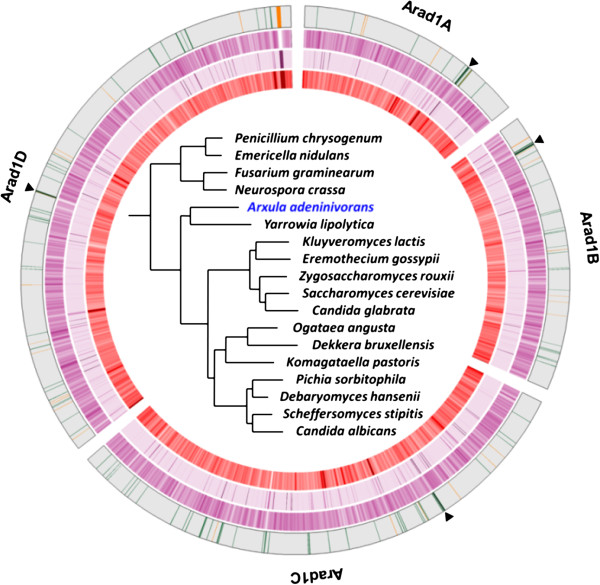
Figure 1.
Circos map of the complete nuclear genome of A. adeninivorans LS3. Chromosome structure (the outermost circle - circle 1): presumed centromeric positions are indicated by black bands and black triangles outside the circle, and tRNA and rRNA genes by green and orange bands, respectively. Genes (circle 2): density of genes in the filtered gene set across the genome, from a gene count per 15 kb sliding window at 5 kb intervals. Repeat content (circle 3): for creating k-mer density ring, k-mers with length = 20 in whole genome using jellyfish program v. 1.1.1 (http://www.cbcb.umd.edu) were counted, a position map of k-mer count was created, k-mers counted in blocks of 3 kb were divided by 3,000 and the data was plotted using Circos’s heatmap. 454 reads mapped to chromosomes (circle 4): density of 454 reads mapped to chromosomes, from a 454 read count per 9 kb sliding window at 3 kb intervals. Underlined blocks indicate alignment in the reverse strand. In the centre of the Circos map the phylogenetic relationship of A. adeninivorans is presented as inferred by gene tree parsimony analysis of the complete A. adeninivorans phylome. k-mer, tuple of length k.