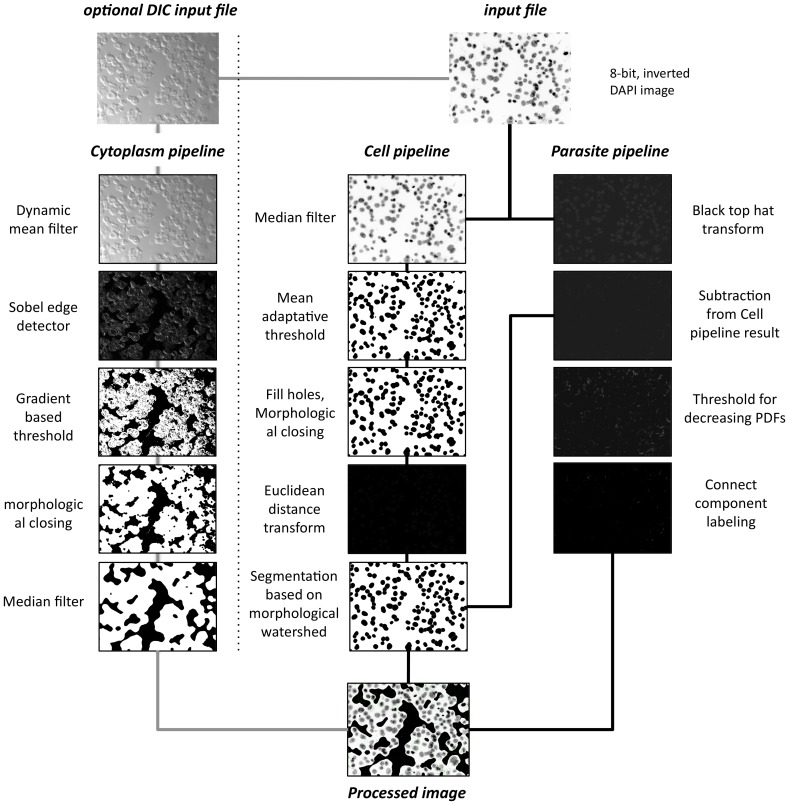
Figure 1. INsPECT processing pipeline. Cell pipeline.
Input image is smoothed using median filter of radius 15. Smoothed image is thresholded using Mean Adaptive Threshold. Window size is assigned automatically and here it is 65. Holes are filled using Binary Fill Holes algorithm and cell scraps are removed using Morphological Closing with structure element radius of 15. Image is transformed to Euclidean Distance domain. Morphological Watershed based segmentation is used to separate overlapped cells and quantify final valid cells. Parasite Pipeline. Top Hat transform is applied to input image to extract tiny details. Cell Pipeline resulted image is subtracted from the current image to filter out tiny pixels belonging to cell portion. Proposed threshold is applied to filter out random noises and other by-products and keep candidates alive for real parasites. Foreground pixels are grouped using Connected Component Labeling algorithm and using minimum and maximum valid sizes taken from user, final valid parasites are extracted from images. Cytoplasm Pipeline. Using Dynamic Mean Filter raw, phase contrast or DIC image is smoothed. Sobel Edge Detector operator is used to extract probable edge candidates for cytoplasm traces. Proposed Cytoplasm threshold is used to discriminate weak from strong edge pixels and obtain probable cytoplasm pixel candidates. Morphological Closing with rectangular structure element of size 15 is applied to fill the holes and Merge Cytoplasm binary regions. Binary Median Filter of radius 25 smoothes cytoplasm regions. Processed Image. Cytoplasm pipeline result will be masked to input DAPI image and then cell and parasites are marked in the image using cell and parasite pipeline results respectively.