Figure 1.
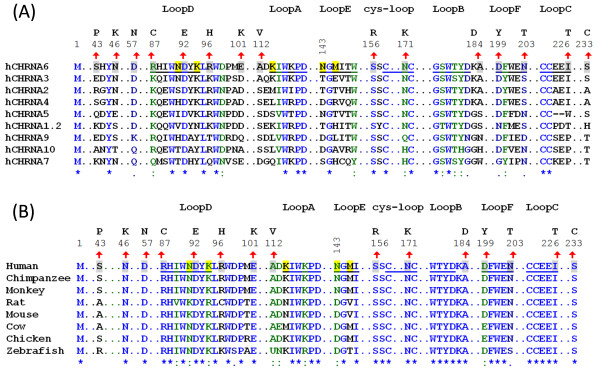
Localizations of N-terminal variations to primary/secondary structure of nAChR hα6 subunits. (A) Degree of conservation of variant residues in nAChR hα6 subunit in relation to other human nAChR α subunits: Human nAChR α subunits (α1-α7, α9 and α10) are aligned using ClustalW. Indicated residues in nAChR hα6 subunits undergoing variation are fully (Asp92 and Ser156), strongly (Arg87 and Asn171) and weakly (Asp57, Asp199 and Asn203) conserved in human nAChR α subunits. Some of the variations in nAChR hα6 subunit are localized to indicated loop regions: Ala112Val (loop A), Ala184Asp (loop B), Ile226Thr (loop C), Asp92Glu (loop D), Asp199Tyr (loop F) and Asn203Thr (loop F) and Asn171Lys (cysteine-loop). (B) Degree of conservation of variant residues in nAChR hα6 subunit in relation to nAChR α6 subunits from other organisms: nAChR α6 protein sequences extracted from (GenBank) NP_067344.2 (Mouse), NP_004189.1 (Human), NP_990695.1 (Chicken), NP_476532.1 (Rat), NP_001029266.1 (Chimpanzee), XP_001099152.1 (Monkey), XP_584902.3 (Cow) and NP_001036149.1 (Zebrafish) are aligned by using ClustalW. For both (A) and (B); numbering begins at translation start methionine of nAChR hα6 subunit and is shown in the regions of interest. However, only segments of the alignment are presented to identify WT nAChR hα6 subunit AA residues (shaded, upward arrow mark and numbers above them) and their corresponding variations (noted above the numberings). Symbols below sequences indicate fully (*), strongly (:) or weakly (.) conserved residues: hα6 subunit AA residues at positions 87 (Arg), 92 (Asp), 156 (Ser) and 171 (Asn) are conserved in both human nAChR α subunits and nAChR α6 subunits of other organisms. Also shown (shaded) are the nAChR hα6 subunit residues including the loop E residue N143 that alone or in combination Met145 influences the function of hα6*-nAChRs [26].
