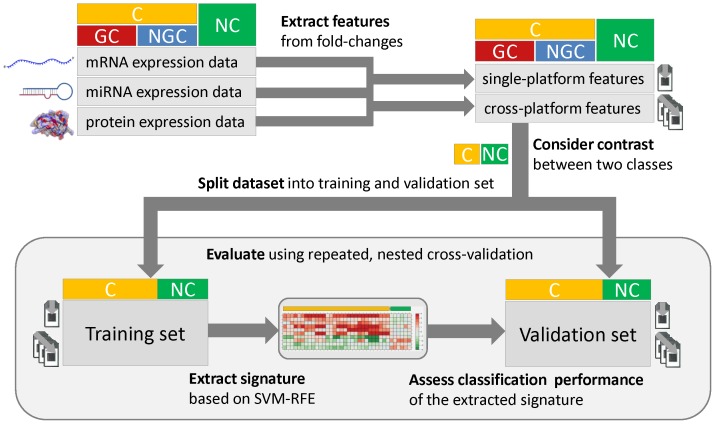
Figure 2. Workflow used for signature extraction and evaluation of classification performance.
For the multi-level omics data available in this study, which include mRNA, miRNA, and protein expression profiles of diverse compounds, fold changes were calculated for each gene and sample that could be confidently assigned to a certain compound class (C: carcinogens, GC: genotoxic carcinogens, NGC: non-genotoxic carcinogens, NC: non-carcinogens). While traditionally used single-platform features simply correspond to fold changes observed on each specific biological level, cross-platform features capture molecular interactions and pathway alterations, which can be inferred by integrating omics data across multiple levels. For each class contrast (e.g., C vs. NC) of interest, the dataset was split into a training set and a validation set. Using the SVM-RFE feature selection technique, a predictive signature for class discrimination was extracted, which was then used to predict the carcinogenic class of the samples in the validation set. By embedding this process into a 2-fold cross-validation with 10 repetitions that use different random splits of the data, the classification performance can be robustly estimated based on the mean area under the ROC curve.