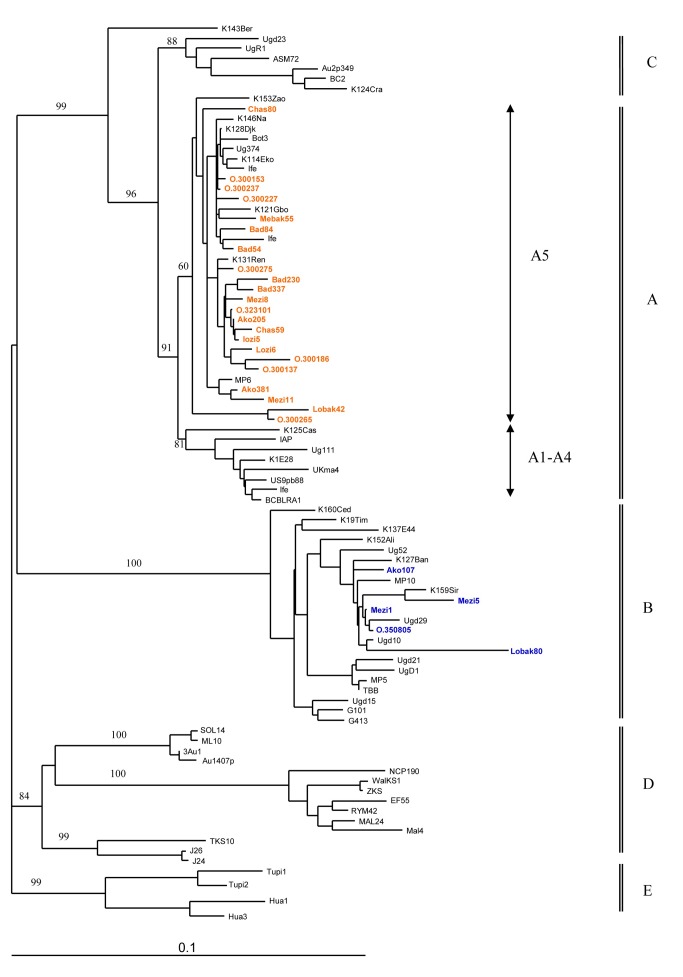
Figure 3. Phylogenetic relationships between the 27 unique new KSHV/HHV-8 sequences.
The phylogenetic tree includes the 29 new 633-types prototypes from healthy persons and KS patients. Amplification was done with primers K1AG75s: GACCTTGTTGGACATCCCGTACAATC, K1AG1200as: AGGCCATGCTGTAAGTAGCACGGTT for the outter fragment and VR1s: ATCCTTGCCAAYATCCTGGTATTGBAA and VR2 as1: AGTACCAMTCCACTGGTTGYGTAT for the inner fragment. Amplified products for 29 samples were directly sequenced. Once the sequences obtained, a multiple sequence alignment was performed with the DAMBE program (v.4.2.13) on the basis of a previous amino acid alignment created from the original sequences. The final alignment was submitted to the Modeltest program (v.3.6) to select the best evolutionary model, according to the Akaike Information Criterion, to apply for the phylogenetic analyses. The phylogeny was derived by both the neighbor-joining (NJ) and maximum parsimony (MP) method, performed in the PAUP program (v.4.0b10) (Sinauer Associates, Sunderland, MA, USA) and the reliability of the inferred tree was evaluated by bootstrap analysis on 1000 replicates. New A5 sequences are shown in bulk red and B sequences are in bulk blue. The tree is drawn to scale with 0.1 nucleotide replacements per site.