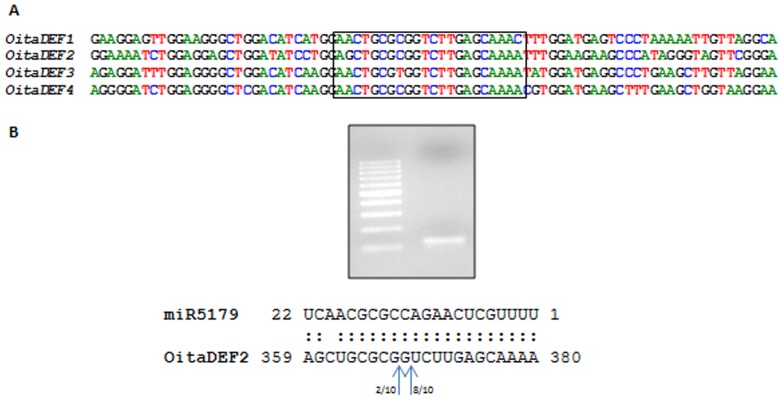
Figure 5. Cleavage of the homolog of miR5179 on OitaDEF2 in O. italica.
A) Nucleotide alignment of the putative target site of miR5179 on OitaDEF2 and its surrounding region with the corresponding region on OitaDEF1, OitaDEF3 and OitaDEF4. The nucleotide sequences span from the position 330 to 410, considering the first nucleotide of the ATG start codon as position 1. B) Agarose gel electrophoresis of the modified 5′ RACE experiment on OitaDEF2 run together with the 100 bp ladder (Fermentas). The alignment of the miR5179 and its target site on OitaDEF4 is reported and the arrows indicate the position of the cleavage and the number of clones corresponding to each site as deduced by the cloning and sequencing of the obtained fragment (see Methods).