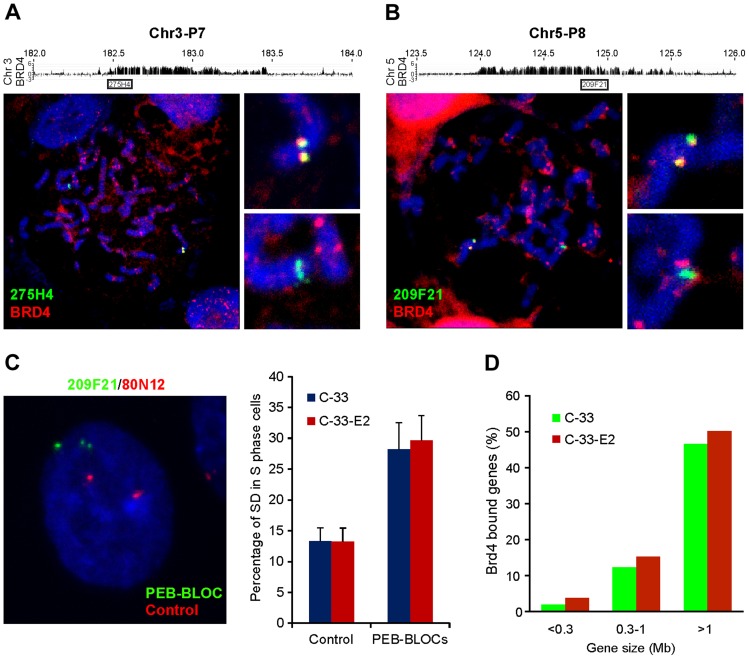
Figure 5. PEB-BLOCs share properties with fragile sites and are targets for chromosomal deletion.
A, B. Chromosomes were spread from mitotic C-33-1E2 cells induced for E2 expression. BRD4 speckles were identified by immunofluorescence, fixed, and hybridized with PEB-BLOC specific FISH probes (107E21, 209F21, 274H4, or 365P10). A representative example is shown for probes 275H4 (A, Chr3-P7) and 209F21 (B, Chr5-P8). The BRD4 signal is red and the FISH signal is green. C. C-33-1E2 cells, induced for E2 expression were analyzed with specific FISH probes. PEB-BLOCs (209F21, 90I21, 274H4, 365P10, 1062A20; green) and control regions (80N12 and 379K17; red). Approximately 50 S-phase cells were analyzed for the SD pattern (one single, one double dot) FISH signals for each PEB-BLOCs or control region. This SD pattern represents asynchronous replication at the allele being examined. Average values and STDEV were calculated for two independent experiments on two control regions (80N12 and 379K17), and 4 PEB-BLOCs (Chr3-P7, Chr5-P8, Chr10-P4, Chr12-P3). D. Correlation between gene size and overlapping PEB-BLOC. The overlap was calculated for a region encompassing the gene body plus 5 kb upstream.