FIGURE 4.
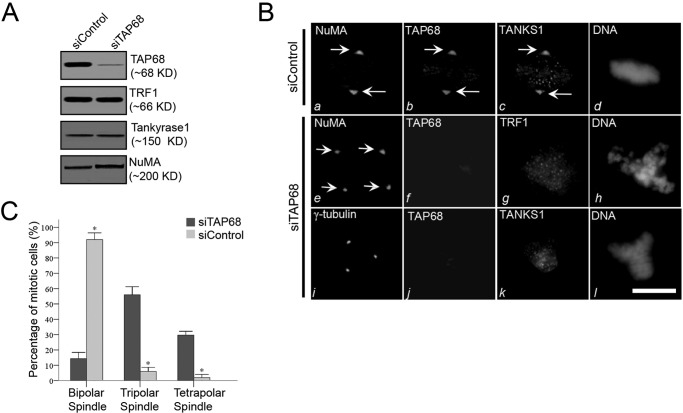
TAP68 mediates TRF1 translocation to the centrosome and mitotic regulation. A, efficient siRNA-mediated suppression of TAP68. HeLa cells were transfected with a siRNA duplex specific for TAP68 and a scrambled control siRNA duplex. After 48 h, HeLa cells were then harvested for Western blot analysis using antibodies specific for TAP68, TRF1, tankyrase 1 (TANKS1), and NuMA. The analyses indicate the specificity and efficiency of the targeted protein suppression. B, representative immunofluorescence images of HeLa cells transfected with control or TAP68-specific siRNA duplex. 48 h post-transfection, cells were stained for the indicated antibodies and DNA, respectively. In the control duplex-transfected cells, NuMA, TAP68, and tankyrase 1 are all co-localized to the centrosome (panels a–c). In TAP68 siRNA duplex-treated cells, TAP68 protein expression was suppressed (panels f and j), and localization of TRF1 to the centrosome was diminished, although TRF1 distribution on chromosome remained (panel g). The distribution of tankyrase 1 to the centrosome was also eliminated in TAP68-depleted cells (panel k). However, the spindle pole localization of NuMA was not altered by the loss of TAP68 (panel e). Tetrapolar spindle in TAP68-depleted cells was revealed by NuMA staining (panel e). Bars, 10 μm. C, quantitative analyses of the mitotic spindle after TAP68 knockdown. HeLa cells were transfected with TAP68 siRNA oligonucleotide and control oligonucleotide for 36–48 h followed by fixation and staining for DNA, TAP68, and NuMA. After examination under a fluorescence microscope, the centrosome instability phenotypes were categorized as bipolar, tripolar, or tetrapolar spindles. Data represent the percentage of total mitotic cells evaluated. An average of 100 cells from three separate experiments was counted. * indicates a significant difference between the TAP68 siRNA- and control siRNA-transfected cells.