TABLE 1.
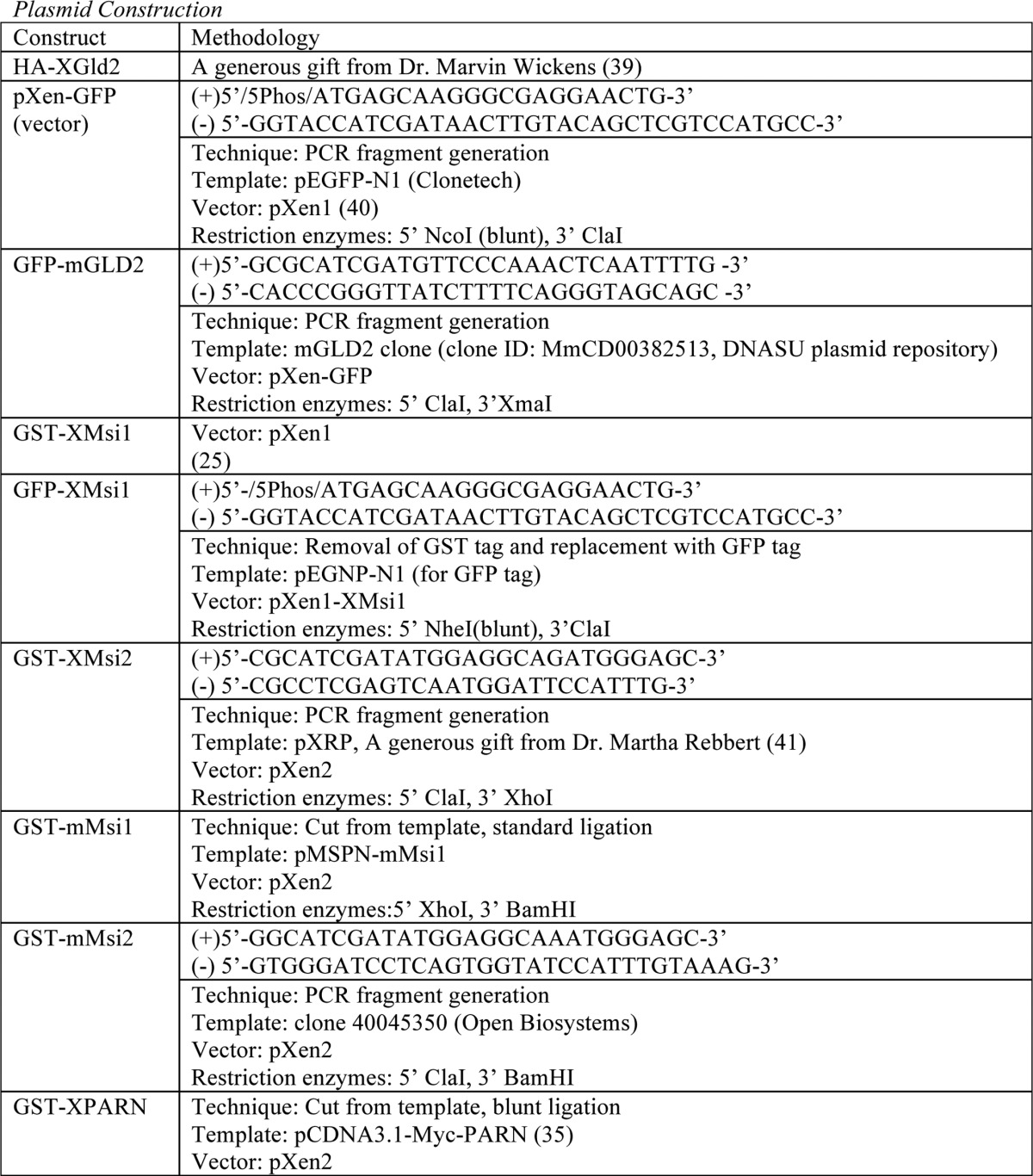
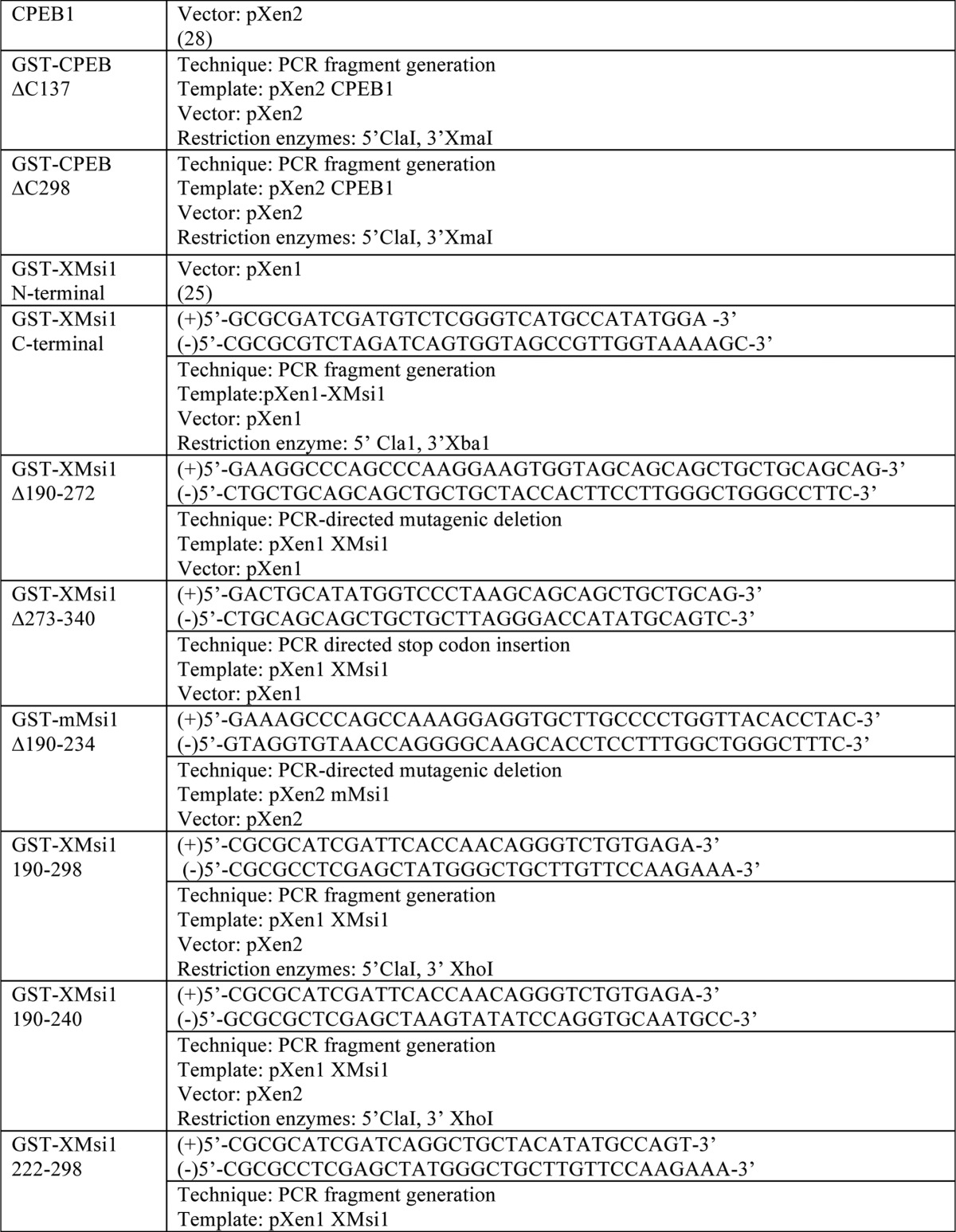
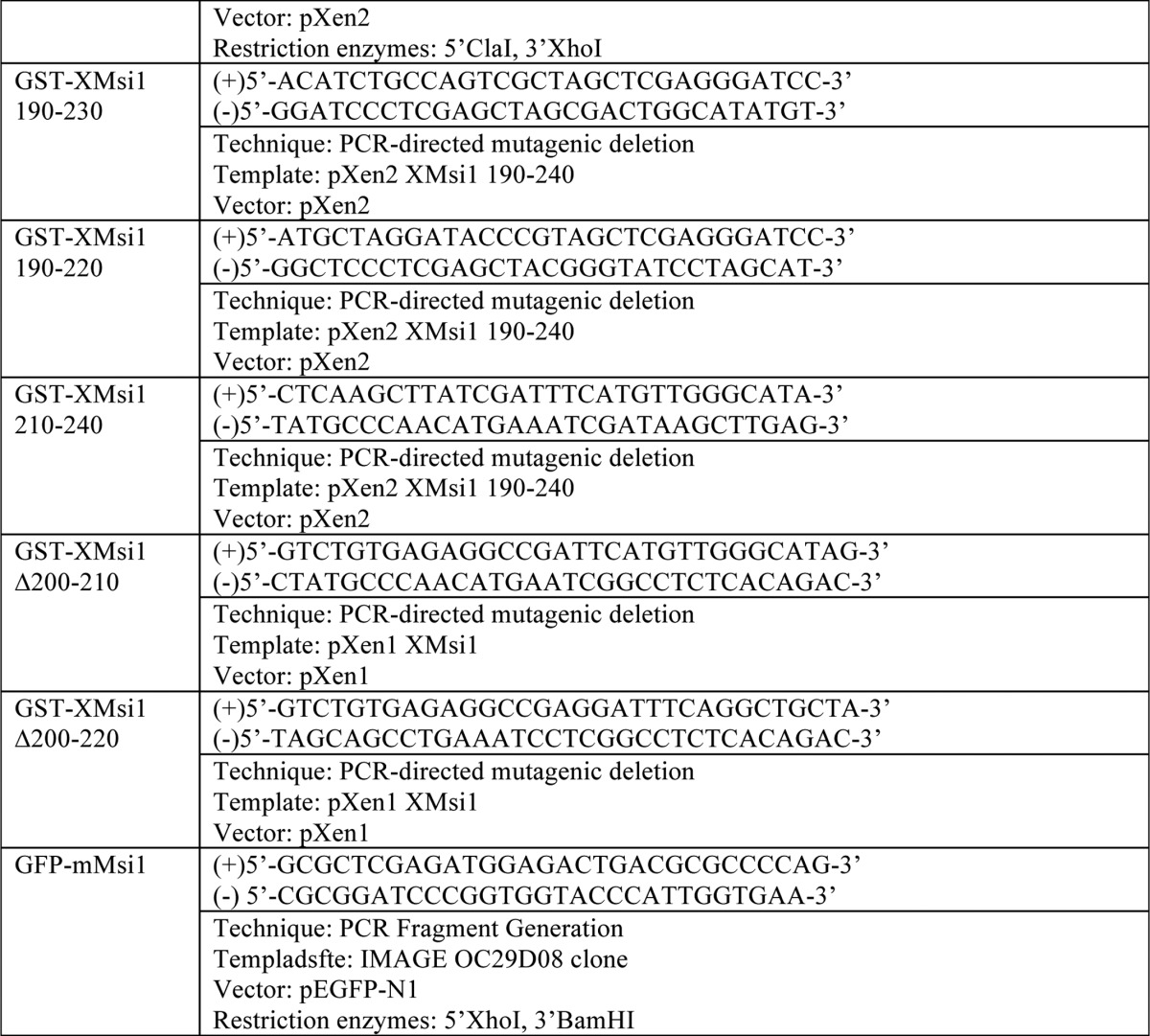
Plasmid construction
For PCR fragment generation, the template was subjected to PCR amplification using the indicated primers. Resulting PCR fragments were then purified using agarose gel electrophoresis followed by cleanup using a QIAquick gel extraction kit (Qiagen). Next, the fragments and destination vector were digested using the indicated restriction enzymes and again purified and cleaned up using gel electrophoresis and the QIAquick kit. The fragment and vector were then ligated using the T7 Quick Ligase (New England Biolabs). Finally, the ligated fragment/vector was used to transform competent DH5-α Escherichia coli. For PCR-directed mutagenic deletion, the template was subjected to PCR amplification of the entire plasmid. Primer sequence “looped out” the desired sequence for deletion. PCR-directed stop codon insertion is the same as the PCR-directed mutagenic deletion, except primers directed insertion of a stop codon rather than deletion.