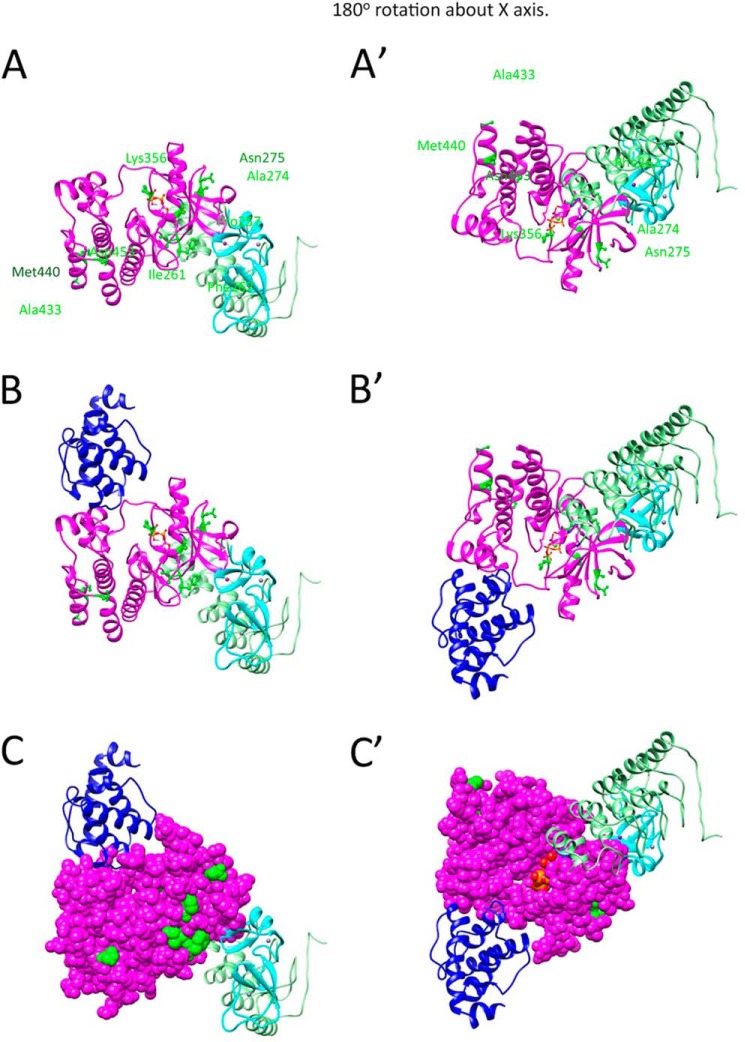
FIGURE 7.
Residues involved in PAT-4 suppressor mutations on the surface of a homology model of PAT-4 are unobstructed after PAT-4 has bound to α-parvin (PAT-6) and to an ankyrin-LIM domain complex. A, two views of a model of PAT-4 (ILK) (magenta) with ankyrin repeats of ILK (light green) and PINCH LIM1 (light blue). These models were generated by substituting the ankyrin-PINCH LIM1 structure from 3F6Q (33) for the inhibitor in the human CDK6/p19INK4D structure (34). Note that ankyrin repeats-PINCH LIM1 complex does not overlap or cover the sites of the PAT-4 suppressor mutations. However, the LIM1 domain of PINCH is near the suppressor mutation Phe-262. B, two views of the composite model: PAT-4 (magenta), parvin (3KMW, dark blue), ankyrin repeats (3F6Q, light green) and PINCH LIM1 (3F6Q, light blue). Note that ankyrin-PINCH LIM1 binds on the opposite surface of PAT-4 as parvin. C, two views of the same complex shown in B, with PAT-4 in space fill mode. The suppressor mutations are shown in green. Note that a majority of the suppressor mutations (Pro-257, Ile-261, Phe-262, Ala-274, Asn-275, Lys-356, and Asn-453) are on the surface between the parvin and PINCH LIM1, on the face opposite the ankyrin repeats.