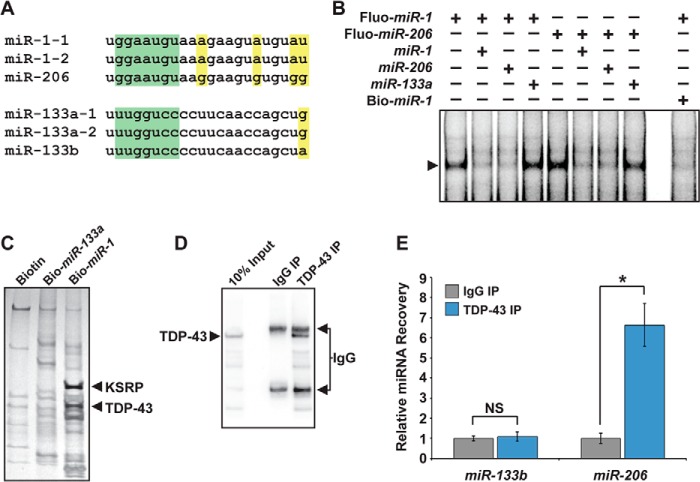
FIGURE 1.
TDP-43 interacts with the miR-1/miR-206 family, but not miR-133. A, sequence alignment of the miR-1/miR-206 family or the miR-133 family. miRNA seed sequence is highlighted in green. Residues that differ among family members are highlighted in yellow. B, EMSAs revealed an miRNA-protein complex (arrow) in undifferentiated C2C12 cell lysate that interacted with fluorescently labeled miR-1 or miR-206 probe (Fluo-miR-1, Fluo-miR-206). These interactions could be competed with unlabeled miR-1 or miR-206, but not with unlabeled miR-133a. 5′-biotinylated miR-1 (Bio-miR-1) also effectively competed for binding. C, eluates from negative control (Biotin), Bio-miR-133a, or Bio-miR-1 pulldowns were run on denaturing gels. Proteins enriched in the Bio-miR-1 pulldown lane were identified by mass spectrometry and included KSRP and TDP-43. D, Western analysis detecting TDP-43 protein following cross-linking and immunoprecipitation from C2C12 skeletal myoblasts using α-TDP-43 (TDP-43 IP), rabbit IgG (IgG IP), or 10% input showed efficient and specific recovery of TDP-43 protein. E, qRT-PCR to detect miR-206 and miR-133b after TDP-43 cross-linking and immunoprecipitation performed in D revealed preferential interaction of TDP-43 with miR-206 versus miR-133b. Results were normalized to pulldown with IgG (NS, not significant; *, p < 0.05).