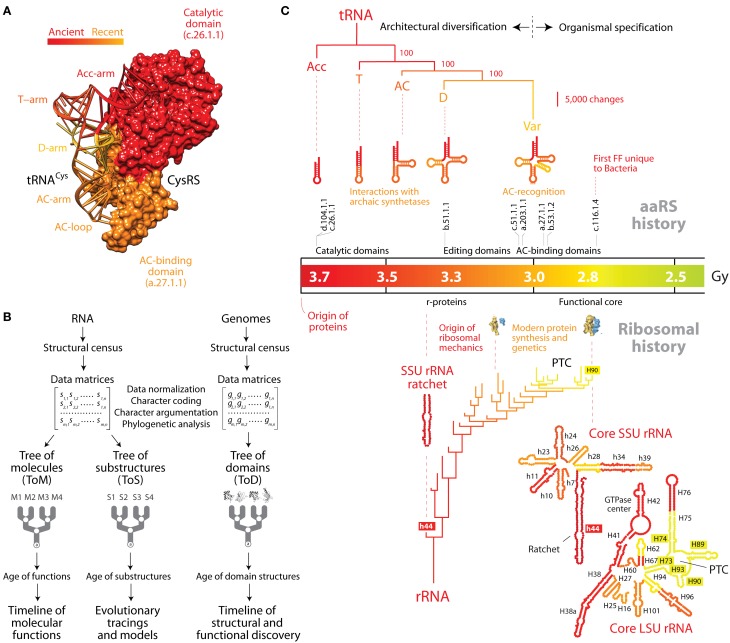
Figure 1.
The natural history of tRNA inferred from nucleic acid-protein interactions and structural phylogenomics. (A) The history of tRNA portrays the history of its interactions with cognate aminoacyl-tRNA synthetase (aaRS) protein enzymes. This is exemplified by the domains of the tRNA and cysteinyl-tRNA synthetase binary complex (PDB entry 1U0B), which are colored according to their age. The ancient “top half” of tRNA embeds a “operational code” in the identity elements of the acceptor arm that interact with the catalytic domain of aaRSs through classes I and II modes of tRNA recognition. The evolutionarily recent “bottom half” of tRNA holds the standard code in identity elements of the anticodon loop that interact with anticodon-binding domains of aaRSs. (B) Flow diagram showing the retrodiction strategy used to build phylogenetic trees of RNA molecules (ToMs) and associated trees of substructures (ToSs), and trees of protein domains (ToDs). The structures of RNA molecules are first decomposed into substructures. Structural features of substructures such as helical stem tracts and unpaired regions are coded as phylogenetic characters and assigned character states according to an evolutionary model that polarizes character transformation toward an increase in conformational order (character argumentation). Coded characters (s) are arranged in data matrices, which can be transposed. Phylogenetic analysis using maximum parsimony optimality criteria generates rooted ToMs and ToSs. A census of domain structures in proteomes of hundreds of completely sequenced organisms is used to compose data matrices, which are then used to build ToDs. Elements of the matrix (g) represent genomic abundances of domain structures in proteomes, defined at different levels of classification of domain structure (e.g., SCOP folds, superfamilies, and families). They are converted into multi-state phylogenetic characters with character states transforming according to linearly ordered and reversible pathways. Embedded in the trees of nucleic acids and proteins are timelines that assign age to molecular structures and associated functions. (C) The natural history of tRNA and rRNA overlap when they are mapped onto a timeline of protein domain history. A tree of tRNA substructures (ToS) was derived from statistical phylogenetic characters that define a molecular morphospace (the Shannon entropy of the base-pairing probability matrix, base-pairing propensity and mean length of stem structures) in 571 tRNA molecules. The optimal most parsimonious tree (43,281 steps; consistency index = 0.853, retention index = 0.654, rescaled consistency index = 0.557, g1 = −1.033) was recovered from a branch-and-bound search. The most basal subtree of a ToS describing the evolution of the rRNA core (Harish and Caetano-Anollés, 2012) is also shown. Both trees are anchored to the geological record via an evolutionary timeline of first appearance of protein domains that are capable of establishing crucial interactions with the RNA molecules (see description in the main text). AC, anticodon; PTC, peptidyl transferase center.