Figure 3. Identification of cell‐cycle related DNA motifs and transcription factors.
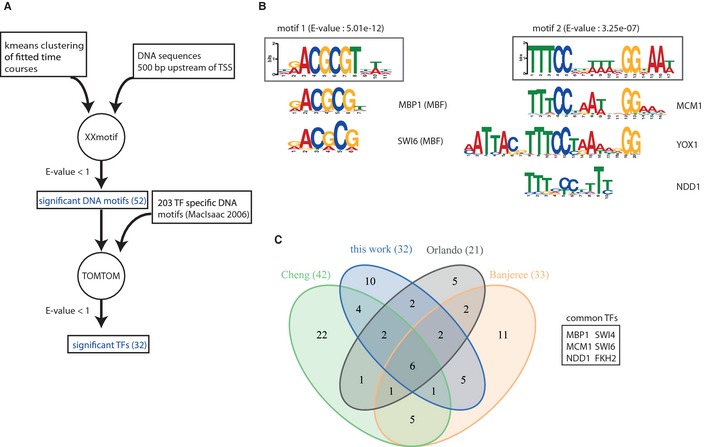
- Workflow, consisting of a motif discovery step using XXmotif and a TF detection step using TOMTOM. The inputs to XXmotif are the 500 bp upstream sequences of sets of co‐regulated genes. The resulting list of significantly enriched motifs is processed by TOMTOM to find TFs with matching binding sites.
- Sequence logos of the two top motifs and their associated TFs (MBF for motif 1; MCM1, YOX1 and NDD1 for motif 2) together with their E‐value.
- Venn diagram showing the overlap of various integrative bioinformatic methods for the prediction of cell‐cycle related TFs.
