Figure 6.
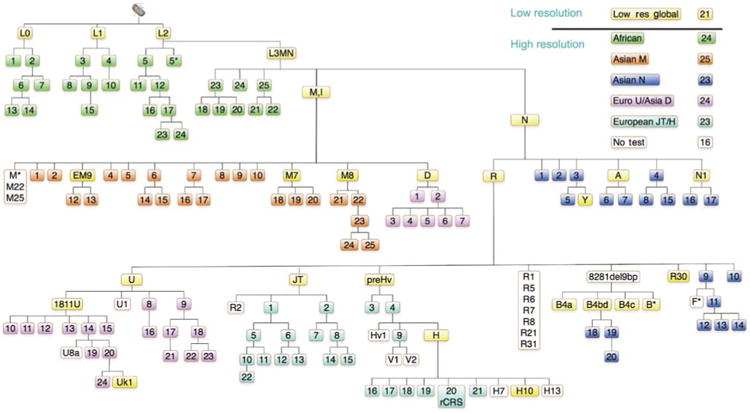
Hierarchical SNV classification of human mtDNA haplogroups.
Aggregates of SNVs are used to define the branches of different portions of the mtDNA tree (Ruiz-Pesini et al., 2007) and are color coded. To classify unknown mtDNAs, they are first divided into major macro-haplogroups and haplogroups using the 21 ‘low resolution’ SNVs, shown in yellow. The mtDNAs for each macro-haplogroup or major haplogroup branch of the tree are then assembled and further subclassified together using the appropriate ‘high resolution’ haplogroup screens. The full version of this tree containing the nucleotide positions and variant nucleotides can be found at the following website: http://www.mitomap.org/pub/MITOMAP/MITOMAPFigures/SNaPshot-Sets.pdf. The SNVs used to define each region of the mtDNA tree are as follows: low resolution global SNVs (yellow): L0(m.13276G), L1(m.13789C), L2(m.7146G), L3MN(m.3594C), M(m.10400T), EM9(m.4491A), M7(m.9824C), M8(m.8584A), D(m.5178A), N(m.9540T), A(m.1736G), R(m.12705C), R30(m.8584A), U(m.12308G), U1811(m.1811G), JT(m.4216C), preHv(m.11719G), H(m.7028C), H10(m.4216C), B4a(m.5465C), B4bd(m.15535T), B4c(m.15346C), B*(m.8584A), N1 (m.10238C). African SNVs (green): 1(m.4232T) 2(m.9818G), 3(m.8027G), 4(m.5046C), 5(m.8206G), 5(m.13590G), 6(m.9755G+m.15431G), 7(m.7257A), 8(m.6917G), 9(m.10321T), 10(m.3693G), 11(m.5069(T)), 12(m.11944T), 13(m.5096A), 14(m.5147G),15(m.6150G), 16(m.7771A), 17(m.7624(A)), 18(m.14905C), 19(m.9554G), 20(m.6221A), 21(m.5147A), 22 (m.6221C) 23(m.4767A), 24(m.15849C). Asian M SNVs (orange): 1(m.12403G), 2(m.15431C), 4(m.15218A), 5(m.8108), 6(m.4833A), 7(m.12940G), 8(m.10986), 9(m.5319T), 10(m.11482), 12(m.7598G), 13(m.3394T), 14(m.15497G), 15(m.13563T), 16(m.14025A), 17(m.11061C), 18(m.4958A), 19(m.7853G), 20(m.5442T), 21(m.9090A), 22(m.14318T), 23(m.15204A), 24(m.12672A), 25(m.3816T). Asian N SNVs (dark blue): 1(m.8404A), 2(m.9140C), 3(m.5417G), 4(m.5460G+8994G), 5(m.12358T), 6(m.8563T), 7(m.8027G), 8(m.3505A), 9(m.3970G+13928C), 10(m.10118A), 11(m.10310C), 12(m.10609T), 13(m.12338T+13708G), 14(m.5263G), 15(m.7864C), 16(m.4529A), 17(m.6221T+14470T), 18(m.11914C), 19(m.4820G), 20(m.9950T). Asian D/European U SNVs (purple): 1(m.5301T), 2(m.3010G+m.8414G), 3(m.2092C), 4(m.3316G), 5(m.14979T), 6(m.9824A), 7(m.10181C), 8(m.3348T), 9(m.9477G), 10(m.13020A), 11(m.15454A), 12(m.5360C), 13(m.15693T), 14(m.9698T), 15(m.6386C), 16(m.7805C), 17(m.14793A), 18(m.14182A), 19(m.14167C), 20(m.14798A), 21(m.15218G), 22(m.13637A), 23(m.5656T), 24(m.9716T). European JT/H SNVs (turquoise): 1(m.13708G), 2(m.4917A), 3(m.13188C), 4(m.14766C), 5(m.3010C), 6(m.15257G), 7(m.12633C), 8(m.14233A), 9(m.4580G), 10(m.14798T), 11(m.5460G+13879A), 12(m.15812G), 13(m.10499T), 14(m.13965A), 15(m.5147G), 16(m.3010C), 17(m.4024A+5004A), 18(m.4336T), 19(m.3915G), 20(m.1438A+4769T), 21(m.6776T), 22(m.3394).
