FIG 3.
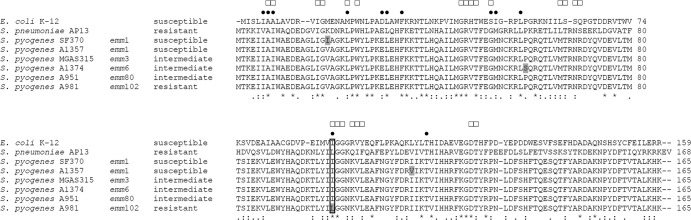
Comparison of amino acid sequences of the intrinsic DHFR of trimethoprim-susceptible E. coli K-12, trimethoprim-susceptible S. pyogenes SF370 and A1357, trimethoprim-intermediate S. pyogenes MGAS315, A1374, and A951, and trimethoprim-resistant S. pyogenes A981 and S. pneumoniae AP13 (12). Isolate designations, emm types of S. pyogenes isolates, and resistance classifications are given on the left. Indicated are amino acid positions involved in trimethoprim (●) and NADPH cofactor (□) binding of the E. coli K-12 enzyme (13–16). Differences between the S. pyogenes DHFRs are highlighted in gray. Identical amino acids (*), conserved substitutions (:), and semiconserved substitutions (.) are indicated as determined by multiple alignment of all sequences. Amino acid positions are indicated on the right. A box indicates position 100.
